Specialized Plastids Trigger Tissue-Specific Signaling for Systemic Stress Response in Plants
- PMID: 30135097
- PMCID: PMC6181059
- DOI: 10.1104/pp.18.00804
Specialized Plastids Trigger Tissue-Specific Signaling for Systemic Stress Response in Plants
Abstract
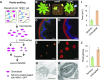
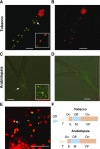
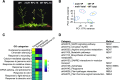
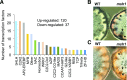
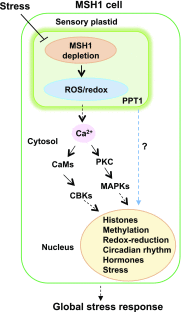
Plastids comprise a complex set of organelles in plants that can undergo distinctive patterns of differentiation and redifferentiation during their lifespan. Plastids localized to the epidermis and vascular parenchyma are distinctive in size, structural features, and functions. These plastids are termed "sensory" plastids, and here we show their proteome to be distinct from chloroplasts, with specialized stress-associated features. The distinctive sensory plastid proteome in Arabidopsis (Arabidopsis thaliana) derives from spatiotemporal regulation of nuclear genes encoding plastid-targeted proteins. Perturbation caused by depletion of the sensory plastid-specific protein MutS HOMOLOG1 conditioned local, programmed changes in gene networks controlling chromatin, stress-related phytohormone, and circadian clock behavior and producing a global, systemic stress response in the plant. We posit that the sensory plastid participates in sensing environmental stress, integrating this sensory function with epigenetic and gene expression circuitry to condition heritable stress memory.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures


References
-
- Abdelnoor RV, Christensen AC, Mohammed S, Munoz-Castillo B, Moriyama H, Mackenzie SA (2006) Mitochondrial genome dynamics in plants and animals: convergent gene fusions of a MutS homologue. J Mol Evol 63: 165–173 - PubMed
-
- Barsan C, Zouine M, Maza E, Bian W, Egea I, Rossignol M, Bouyssie D, Pichereaux C, Purgatto E, Bouzayen M, et al. (2012) Proteomic analysis of chloroplast-to-chromoplast transition in tomato reveals metabolic shifts coupled with disrupted thylakoid biogenesis machinery and elevated energy-production components. Plant Physiol 160: 708–725 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

