Cottontail Rabbit Papillomavirus E1 and E2 Proteins Mutually Influence Their Subcellular Localizations
- PMID: 30135125
- PMCID: PMC6189495
- DOI: 10.1128/JVI.00704-18
Cottontail Rabbit Papillomavirus E1 and E2 Proteins Mutually Influence Their Subcellular Localizations
Abstract
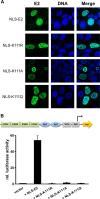
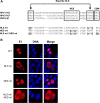
The papillomavirus (PV) E2 protein is a nuclear, sequence-specific DNA-binding protein that regulates transcription and nuclear retention of viral genomes. E2 also interacts with the viral E1 protein to replicate the viral genome. E2 residue K111 is highly conserved among PV and has been implicated in contributing to nuclear transport, transcription, and replication. Cottontail rabbit (Sylvilagus floridanus) PV (CRPV or SfPV1) E2 K111R, A, or Q mutations are transcription deficient and localized to the cytoplasm, comparable to other PV types. The addition of a nuclear localization signal (NLS) resulted in nuclear E2 K111 mutant proteins but did not restore transcriptional activation, and this is most likely due to an impaired binding to the cellular Brd4 protein. Surprisingly, coexpression of E1 with E2 K111 mutations resulted in their nuclear localization and, for K111A and R mutations, the activation of an E1/E2-dependent reporter construct. Interestingly, the nuclear localization of E2 K111Q mutant protein was independent from the presence of the conserved bipartite NLS in E1 and the direct interaction between E1 and E2. On the other hand, the cytoplasmic E1 NLS mutation could be targeted to the nucleus by wild-type E2, and this was dependent upon an interaction between E1 and E2. In summary, our studies have uncovered that E1 and E2 control each other's subcellular localization: direct binding of E2 to E1 can direct E1 to the nucleus independently from the E1 NLS, and E1 can direct E2 to the nucleus without an intact NLS or direct binding to E2.IMPORTANCE Papillomaviruses encode the DNA-binding E1 and E2 proteins, which form a complex and are essential for genome replication. Both proteins are targeted to the nucleus via nuclear localization signals. Our studies have uncovered that cytoplasmic mutant E1 or E2 proteins can be localized to the nucleus when E1 or E2 is also present. An interaction between E1 and E2 is necessary to target cytoplasmic E1 mutant proteins to the nucleus, but cytoplasmic E2 mutant proteins can be targeted to the nucleus without a direct interaction, which points to a novel function of E1.
Keywords: E1; E2; nuclear localization; papillomavirus; replication.
Copyright © 2018 American Society for Microbiology.
Figures








References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

