Phosphoserine acidic cluster motifs bind distinct basic regions on the μ subunits of clathrin adaptor protein complexes
- PMID: 30135209
- PMCID: PMC6177606
- DOI: 10.1074/jbc.RA118.003080
Phosphoserine acidic cluster motifs bind distinct basic regions on the μ subunits of clathrin adaptor protein complexes
Abstract
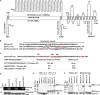
Protein trafficking in the endosomal system involves the recognition of specific signals within the cytoplasmic domains (CDs) of transmembrane proteins by clathrin adaptors. One such signal is the phosphoserine acidic cluster (PSAC), the prototype of which is in the endoprotease furin. How PSACs are recognized by clathrin adaptors has been controversial. We reported previously that HIV-1 Vpu, which modulates cellular immunoreceptors, contains a PSAC that binds to the μ subunits of clathrin adaptor protein (AP) complexes. Here, we show that the CD of furin binds the μ subunits of AP-1 and AP-2 in a phosphorylation-dependent manner. Moreover, we identify a potential PSAC in a cytoplasmic loop of the cellular transmembrane Serinc3, an inhibitor of the infectivity of retroviruses. The two serines within the PSAC of Serinc3 are phosphorylated by casein kinase II and mediate interaction with the μ subunits in vitro The sites of these serines vary among mammals in a manner suggesting host-pathogen conflict, yet the Serinc3 PSAC seems dispensable for anti-HIV activity and for counteraction by HIV-1 Nef. The CDs of Vpu and furin and the PSAC-containing loop of Serinc3 each bind the μ subunit of AP-2 (μ2) with similar affinities, but they appear to utilize different basic regions on μ2. The Serinc3 loop requires a region previously reported to bind the acidic plasma membrane lipid phosphatidylinositol 4,5-bisphosphate. These data suggest that the PSACs within different proteins recognize different basic regions on the μ surface, providing the potential to inhibit the activity of viral proteins without necessarily affecting cellular protein trafficking.
Keywords: HIV-1 Vpu; Serinc3; acidic cluster; adaptor protein; clathrin; furin; host-pathogen interaction; human immunodeficiency virus (HIV); kinetics; medium subunit; membrane trafficking; phosphorylation; phosphoserine; protein chemistry; protein complex.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures





Similar articles
-
Endocytic activity of HIV-1 Vpu: Phosphoserine-dependent interactions with clathrin adaptors.Traffic. 2017 Aug;18(8):545-561. doi: 10.1111/tra.12495. Epub 2017 Jun 27. Traffic. 2017. PMID: 28504462 Free PMC article.
-
Structural basis of HIV-1 Vpu-mediated BST2 antagonism via hijacking of the clathrin adaptor protein complex 1.Elife. 2014 Apr 29;3:e02362. doi: 10.7554/eLife.02362. Elife. 2014. PMID: 24843023 Free PMC article.
-
Contribution of the clathrin adaptor AP-1 subunit µ1 to acidic cluster protein sorting.J Cell Biol. 2017 Sep 4;216(9):2927-2943. doi: 10.1083/jcb.201602058. Epub 2017 Jul 25. J Cell Biol. 2017. PMID: 28743825 Free PMC article.
-
Conformational regulation of AP1 and AP2 clathrin adaptor complexes.Traffic. 2019 Oct;20(10):741-751. doi: 10.1111/tra.12677. Epub 2019 Aug 6. Traffic. 2019. PMID: 31313456 Free PMC article. Review.
-
HIV-1 accessory proteins--ensuring viral survival in a hostile environment.Cell Host Microbe. 2008 Jun 12;3(6):388-98. doi: 10.1016/j.chom.2008.04.008. Cell Host Microbe. 2008. PMID: 18541215 Review.
Cited by
-
HPV-16 E7 Interacts with the Endocytic Machinery via the AP2 Adaptor μ2 Subunit.mBio. 2022 Dec 20;13(6):e0230222. doi: 10.1128/mbio.02302-22. Epub 2022 Oct 18. mBio. 2022. PMID: 36255238 Free PMC article.
-
Endosomal chloride/proton exchangers need inhibitory TMEM9 β-subunits for regulation and prevention of disease-causing overactivity.Nat Commun. 2025 Apr 1;16(1):3117. doi: 10.1038/s41467-025-58546-3. Nat Commun. 2025. PMID: 40169677 Free PMC article.
-
Plasma Membrane-Associated Restriction Factors and Their Counteraction by HIV-1 Accessory Proteins.Cells. 2019 Sep 2;8(9):1020. doi: 10.3390/cells8091020. Cells. 2019. PMID: 31480747 Free PMC article. Review.
-
Coatopathies: Genetic Disorders of Protein Coats.Annu Rev Cell Dev Biol. 2019 Oct 6;35:131-168. doi: 10.1146/annurev-cellbio-100818-125234. Epub 2019 Aug 9. Annu Rev Cell Dev Biol. 2019. PMID: 31399000 Free PMC article. Review.
-
Mechanism of p38 MAPK-induced EGFR endocytosis and its crosstalk with ligand-induced pathways.J Cell Biol. 2021 Jul 5;220(7):e202102005. doi: 10.1083/jcb.202102005. Epub 2021 May 25. J Cell Biol. 2021. PMID: 34032851 Free PMC article.
References
-
- Voorhees P., Deignan E., van Donselaar E., Humphrey J., Marks M. S., Peters P. J., and Bonifacino J. S. (1995) An acidic sequence within the cytoplasmic domain of furin functions as a determinant of trans-Golgi network localization and internalization from the cell surface. EMBO J. 14, 4961–4975 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

