Prevalence Estimates of Polycystic Kidney and Liver Disease by Population Sequencing
- PMID: 30135240
- PMCID: PMC6171271
- DOI: 10.1681/ASN.2018050493
Prevalence Estimates of Polycystic Kidney and Liver Disease by Population Sequencing
Abstract
Background: Estimating the prevalence of autosomal dominant polycystic kidney disease (ADPKD) is challenging because of age-dependent penetrance and incomplete clinical ascertainment. Early studies estimated the lifetime risk of ADPKD to be about one per 1000 in the general population, whereas recent epidemiologic studies report a point prevalence of three to five cases per 10,000 in the general population.
Methods: To measure the frequency of high-confidence mutations presumed to be causative in ADPKD and autosomal dominant polycystic liver disease (ADPLD) and estimate lifetime ADPKD prevalence, we used two large, population sequencing databases, gnomAD (15,496 whole-genome sequences; 123,136 exome sequences) and BRAVO (62,784 whole-genome sequences). We used stringent criteria for defining rare variants in genes involved in ADPKD (PKD1, PKD2), ADPLD (PRKCSH, SEC63, GANAB, ALG8, SEC61B, LRP5), and potential cystic disease modifiers; evaluated variants for quality and annotation; compared variants with data from an ADPKD mutation database; and used bioinformatic tools to predict pathogenicity.
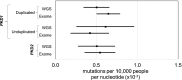
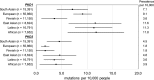
Results: Identification of high-confidence pathogenic mutations in whole-genome sequencing provided a lower boundary for lifetime ADPKD prevalence of 9.3 cases per 10,000 sequenced. Estimates from whole-genome and exome data were similar. Truncating mutations in ADPLD genes and genes of potential relevance as cyst modifiers were found in 20.2 cases and 103.9 cases per 10,000 sequenced, respectively.
Conclusions: Population whole-genome sequencing suggests a higher than expected prevalence of ADPKD-associated mutations. Loss-of-function mutations in ADPLD genes are also more common than expected, suggesting the possibility of unrecognized cases and incomplete penetrance. Substantial rare variation exists in genes with potential for phenotype modification in ADPKD.
Keywords: ADPKD; disease; genetic renal; human genetics; liver cysts; polycystic kidney disease.
Copyright © 2018 by the American Society of Nephrology.
Figures



References
-
- Heyer CM, Sundsbak JL, Abebe KZ, Chapman AB, Torres VE, Grantham JJ, et al.: HALT PKD and CRISP Investigators : Predicted mutation strength of nontruncating PKD1 mutations aids genotype-phenotype correlations in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 27: 2872–2884, 2016 - PMC - PubMed
-
- Song X, Haghighi A, Iliuta I-A, Pei Y: Molecular diagnosis of autosomal dominant polycystic kidney disease. Expert Rev Mol Diagn 17: 885–895, 2017 - PubMed
-
- Harris PC, Bae KT, Rossetti S, Torres VE, Grantham JJ, Chapman AB, et al.: Cyst number but not the rate of cystic growth is associated with the mutated gene in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 17: 3013–3019, 2006 - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

