Ndr kinases regulate retinal interneuron proliferation and homeostasis
- PMID: 30135513
- PMCID: PMC6105603
- DOI: 10.1038/s41598-018-30492-9
Ndr kinases regulate retinal interneuron proliferation and homeostasis
Abstract
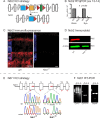
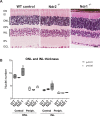
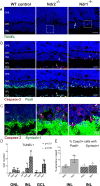
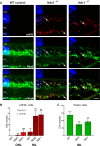
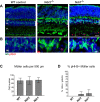
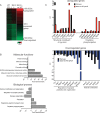
Ndr2/Stk38l encodes a protein kinase associated with the Hippo tumor suppressor pathway and is mutated in a naturally-occurring canine early retinal degeneration (erd). To elucidate the retinal functions of Ndr2 and its paralog Ndr1/Stk38, we generated Ndr1 and Ndr2 single knockout mice. Although retinal lamination appeared normal in these mice, Ndr deletion caused a subset of Pax6-positive amacrine cells to proliferate in differentiated retinas, while concurrently decreasing the number of GABAergic, HuD and Pax6-positive amacrine cells. Retinal transcriptome analyses revealed that Ndr2 deletion increased expression of neuronal stress genes and decreased expression of synaptic organization genes. Consistent with the latter, Ndr deletion dramatically reduced levels of Aak1, an Ndr substrate that regulates vesicle trafficking. Our findings indicate that Ndr kinases are important regulators of amacrine and photoreceptor cells and suggest that Ndr kinases inhibit the proliferation of a subset of terminally differentiated cells and modulate interneuron synapse function via Aak1.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

