Structure of paused transcription complex Pol II-DSIF-NELF
- PMID: 30135580
- PMCID: PMC6245578
- DOI: 10.1038/s41586-018-0442-2
Structure of paused transcription complex Pol II-DSIF-NELF
Abstract
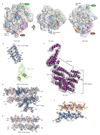
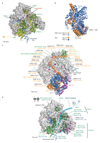
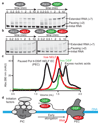
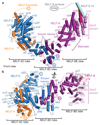
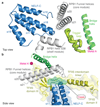
Metazoan gene regulation often involves the pausing of RNA polymerase II (Pol II) in the promoter-proximal region. Paused Pol II is stabilized by the protein complexes DRB sensitivity-inducing factor (DSIF) and negative elongation factor (NELF). Here we report the cryo-electron microscopy structure of a paused transcription elongation complex containing Sus scrofa Pol II and Homo sapiens DSIF and NELF at 3.2 Å resolution. The structure reveals a tilted DNA-RNA hybrid that impairs binding of the nucleoside triphosphate substrate. NELF binds the polymerase funnel, bridges two mobile polymerase modules, and contacts the trigger loop, thereby restraining Pol II mobility that is required for pause release. NELF prevents binding of the anti-pausing transcription elongation factor IIS (TFIIS). Additionally, NELF possesses two flexible 'tentacles' that can contact DSIF and exiting RNA. These results define the paused state of Pol II and provide the molecular basis for understanding the function of NELF during promoter-proximal gene regulation.
Conflict of interest statement
The author declare that they have no competing financial interest.
Figures





Comment in
-
Transcriptional speed bumps revealed in high resolution.Nature. 2018 Aug;560(7720):560-561. doi: 10.1038/d41586-018-05971-8. Nature. 2018. PMID: 30143755 No abstract available.
Similar articles
-
Structure of activated transcription complex Pol II-DSIF-PAF-SPT6.Nature. 2018 Aug;560(7720):607-612. doi: 10.1038/s41586-018-0440-4. Epub 2018 Aug 22. Nature. 2018. PMID: 30135578
-
Distinct negative elongation factor conformations regulate RNA polymerase II promoter-proximal pausing.Mol Cell. 2024 Apr 4;84(7):1243-1256.e5. doi: 10.1016/j.molcel.2024.01.023. Epub 2024 Feb 23. Mol Cell. 2024. PMID: 38401543 Free PMC article.
-
Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex.Proc Natl Acad Sci U S A. 2010 Jun 22;107(25):11301-6. doi: 10.1073/pnas.1000681107. Epub 2010 Jun 4. Proc Natl Acad Sci U S A. 2010. PMID: 20534440 Free PMC article.
-
Pause Patrol: Negative Elongation Factor's Role in Promoter-Proximal Pausing and Beyond.J Mol Biol. 2025 Jan 1;437(1):168779. doi: 10.1016/j.jmb.2024.168779. Epub 2024 Sep 4. J Mol Biol. 2025. PMID: 39241983 Review.
-
Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond.Biochim Biophys Acta. 2013 Jan;1829(1):98-104. doi: 10.1016/j.bbagrm.2012.11.007. Epub 2012 Nov 29. Biochim Biophys Acta. 2013. PMID: 23202475 Review.
Cited by
-
Structures of mammalian RNA polymerase II pre-initiation complexes.Nature. 2021 Jun;594(7861):124-128. doi: 10.1038/s41586-021-03554-8. Epub 2021 Apr 26. Nature. 2021. PMID: 33902107
-
Addressing Transcriptional Dysregulation in Cancer through CDK9 Inhibition.Biochemistry. 2023 Mar 21;62(6):1114-1123. doi: 10.1021/acs.biochem.2c00609. Epub 2023 Feb 28. Biochemistry. 2023. PMID: 36854448 Free PMC article.
-
Unexpected diversity in eukaryotic transcription revealed by the retrotransposon hotspot family of Trypanosoma brucei.Nucleic Acids Res. 2019 Feb 28;47(4):1725-1739. doi: 10.1093/nar/gky1255. Nucleic Acids Res. 2019. PMID: 30544263 Free PMC article.
-
m6A RNA modification in transcription regulation.Transcription. 2021 Oct;12(5):266-276. doi: 10.1080/21541264.2022.2057177. Epub 2022 Apr 5. Transcription. 2021. PMID: 35380917 Free PMC article. Review.
-
Widespread Backtracking by RNA Pol II Is a Major Effector of Gene Activation, 5' Pause Release, Termination, and Transcription Elongation Rate.Mol Cell. 2019 Jan 3;73(1):107-118.e4. doi: 10.1016/j.molcel.2018.10.031. Epub 2018 Nov 29. Mol Cell. 2019. PMID: 30503775 Free PMC article.
References
-
- Shilatifard A, Conaway RC, Conaway JW. The RNA Polymerase II Elongation Complex. Annu Rev Biochem. 2003;72:693–715. - PubMed
-
- Bentley DL, Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986;321:702–706. - PubMed
-
- Kao S-Y, Calman AF, Luciw PA, Peterlin BM. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

