RAP2 mediates mechanoresponses of the Hippo pathway
- PMID: 30135582
- PMCID: PMC6128698
- DOI: 10.1038/s41586-018-0444-0
RAP2 mediates mechanoresponses of the Hippo pathway
Abstract
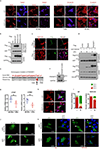
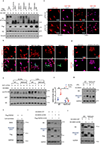
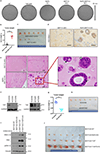
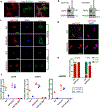
Mammalian cells are surrounded by neighbouring cells and extracellular matrix (ECM), which provide cells with structural support and mechanical cues that influence diverse biological processes1. The Hippo pathway effectors YAP (also known as YAP1) and TAZ (also known as WWTR1) are regulated by mechanical cues and mediate cellular responses to ECM stiffness2,3. Here we identified the Ras-related GTPase RAP2 as a key intracellular signal transducer that relays ECM rigidity signals to control mechanosensitive cellular activities through YAP and TAZ. RAP2 is activated by low ECM stiffness, and deletion of RAP2 blocks the regulation of YAP and TAZ by stiffness signals and promotes aberrant cell growth. Mechanistically, matrix stiffness acts through phospholipase Cγ1 (PLCγ1) to influence levels of phosphatidylinositol 4,5-bisphosphate and phosphatidic acid, which activates RAP2 through PDZGEF1 and PDZGEF2 (also known as RAPGEF2 and RAPGEF6). At low stiffness, active RAP2 binds to and stimulates MAP4K4, MAP4K6, MAP4K7 and ARHGAP29, resulting in activation of LATS1 and LATS2 and inhibition of YAP and TAZ. RAP2, YAP and TAZ have pivotal roles in mechanoregulated transcription, as deletion of YAP and TAZ abolishes the ECM stiffness-responsive transcriptome. Our findings show that RAP2 is a molecular switch in mechanotransduction, thereby defining a mechanosignalling pathway from ECM stiffness to the nucleus.
Conflict of interest statement
Competing interests
Dr. Kun-Liang Guan is a co-founder of Vivace Therapeutics.
Figures










Comment in
-
Rapping about Mechanotransduction.Dev Cell. 2018 Sep 24;46(6):678-679. doi: 10.1016/j.devcel.2018.09.007. Dev Cell. 2018. PMID: 30253168
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

