Be on Target: Strategies of Targeting Alternative and Lectin Pathway Components in Complement-Mediated Diseases
- PMID: 30135690
- PMCID: PMC6092519
- DOI: 10.3389/fimmu.2018.01851
Be on Target: Strategies of Targeting Alternative and Lectin Pathway Components in Complement-Mediated Diseases
Abstract
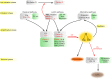
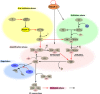
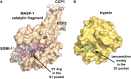
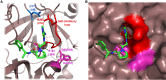
The complement system has moved into the focus of drug development efforts in the last decade, since its inappropriate or uncontrolled activation has been recognized in many diseases. Some of them are primarily complement-mediated rare diseases, such as paroxysmal nocturnal hemoglobinuria, C3 glomerulonephritis, and atypical hemolytic uremic syndrome. Complement also plays a role in various multifactorial diseases that affect millions of people worldwide, such as ischemia reperfusion injury (myocardial infarction, stroke), age-related macular degeneration, and several neurodegenerative disorders. In this review, we summarize the potential advantages of targeting various complement proteins with special emphasis on the components of the lectin (LP) and the alternative pathways (AP). The serine proteases (MASP-1/2/3, factor D, factor B), which are responsible for the activation of the cascade, are straightforward targets of inhibition, but the pattern recognition molecules (mannose-binding lectin, other collectins, and ficolins), the regulatory components (factor H, factor I, properdin), and C3 are also subjects of drug development. Recent discoveries about cross-talks between the LP and AP offer new approaches for clinical intervention. Mannan-binding lectin-associated serine proteases (MASPs) are not just responsible for LP activation, but they are also indispensable for efficient AP activation. Activated MASP-3 has recently been shown to be the enzyme that continuously supplies factor D (FD) for the AP by cleaving pro-factor D (pro-FD). In this aspect, MASP-3 emerges as a novel feasible target for the regulation of AP activity. MASP-1 was shown to be required for AP activity on various surfaces, first of all on LPS of Gram-negative bacteria.
Keywords: alternative pathway; complement inhibitors; complement system; complement-related diseases; lectin pathway.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

