Non-local Parabolic and Hyperbolic Models for Cell Polarisation in Heterogeneous Cancer Cell Populations
- PMID: 30136211
- PMCID: PMC6153854
- DOI: 10.1007/s11538-018-0477-4
Non-local Parabolic and Hyperbolic Models for Cell Polarisation in Heterogeneous Cancer Cell Populations
Abstract
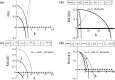
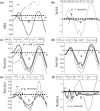
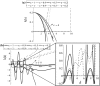
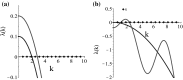
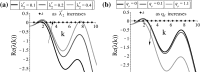
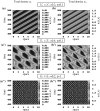
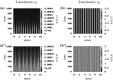
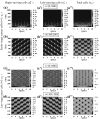
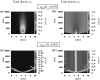
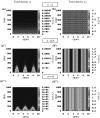
Tumours consist of heterogeneous populations of cells. The sub-populations can have different features, including cell motility, proliferation and metastatic potential. The interactions between clonal sub-populations are complex, from stable coexistence to dominant behaviours. The cell-cell interactions, i.e. attraction, repulsion and alignment, processes critical in cancer invasion and metastasis, can be influenced by the mutation of cancer cells. In this study, we develop a mathematical model describing cancer cell invasion and movement for two polarised cancer cell populations with different levels of mutation. We consider a system of non-local hyperbolic equations that incorporate cell-cell interactions in the speed and the turning behaviour of cancer cells, and take a formal parabolic limit to transform this model into a non-local parabolic model. We then investigate the possibility of aggregations to form, and perform numerical simulations for both hyperbolic and parabolic models, comparing the patterns obtained for these models.
Keywords: Aggregation patterns; Alignment; Cancer cells; Cell–cell interactions; Non-local hyperbolic model; Parabolic limit.
Figures



Similar articles
-
Symmetries and pattern formation in hyperbolic versus parabolic models of self-organised aggregation.J Math Biol. 2015 Oct;71(4):847-81. doi: 10.1007/s00285-014-0842-3. Epub 2014 Oct 15. J Math Biol. 2015. PMID: 25315439
-
Chirality provides a direct fitness advantage and facilitates intermixing in cellular aggregates.PLoS Comput Biol. 2018 Dec 27;14(12):e1006645. doi: 10.1371/journal.pcbi.1006645. eCollection 2018 Dec. PLoS Comput Biol. 2018. PMID: 30589836 Free PMC article.
-
Mathematical modelling of cancer cell invasion of tissue: local and non-local models and the effect of adhesion.J Theor Biol. 2008 Feb 21;250(4):684-704. doi: 10.1016/j.jtbi.2007.10.026. J Theor Biol. 2008. PMID: 18068728
-
Polarity proteins in migration and invasion.Oncogene. 2008 Nov 24;27(55):6970-80. doi: 10.1038/onc.2008.347. Oncogene. 2008. PMID: 19029938 Review.
-
Membrane ruffling of cancer cells: a parameter of tumour cell motility and invasion.Eur J Surg Oncol. 1995 Jun;21(3):307-9. doi: 10.1016/s0748-7983(95)91690-3. Eur J Surg Oncol. 1995. PMID: 7781803 Review.
References
-
- Anderson ARA, Chaplain MAJ, Newman EL, Steele RJC, Thompson AM. Mathematical modelling of tumour invasion and metastasis. J Theor Med. 2000;2(2):129–154. doi: 10.1080/10273660008833042. - DOI
-
- Bitsouni V, Chaplain MAJ, Eftimie R. Mathematical modelling of cancer invasion: the multiple roles of TGF- pathway on tumour proliferation and cell adhesion. Math Models Methods Appl Sci. 2017;27(10):1929–1962. doi: 10.1142/S021820251750035X. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

