Phylogeny Estimation by Integration over Isolation with Migration Models
- PMID: 30137463
- PMCID: PMC6231491
- DOI: 10.1093/molbev/msy162
Phylogeny Estimation by Integration over Isolation with Migration Models
Abstract
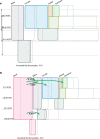
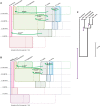
Phylogeny estimation is difficult for closely related populations and species, especially if they have been exchanging genes. We present a hierarchical Bayesian, Markov-chain Monte Carlo method with a state space that includes all possible phylogenies in a full Isolation-with-Migration model framework. The method is based on a new type of genealogy augmentation called a "hidden genealogy" that enables efficient updating of the phylogeny. This is the first likelihood-based method to fully incorporate directional gene flow and genetic drift for estimation of a species or population phylogeny. Application to human hunter-gatherer populations from Africa revealed a clear phylogenetic history, with strong support for gene exchange with an unsampled ghost population, and relatively ancient divergence between a ghost population and modern human populations, consistent with human/archaic divergence. In contrast, a study of five chimpanzee populations reveals a clear phylogeny with several pairs of populations having exchanged DNA, but does not support a history with an unsampled ghost population.
Figures



References
-
- Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F.. 2004. Parallel metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20(3):407–415. - PubMed
-
- Arbogast BS, Edwards SV, Wakeley J, Beerli P, Slowinski JB.. 2002. Estimating divergence times from molecular data on phylogenetic and population genetic timescales. Annu Rev Ecol Syst. 33(1):707–740.
-
- Avise JC. 1994. Molecular markers, natural history and evolution. London: Chapman & Hall.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

