The small GTPase RAB-35 defines a third pathway that is required for the recognition and degradation of apoptotic cells
- PMID: 30138370
- PMCID: PMC6107108
- DOI: 10.1371/journal.pgen.1007558
The small GTPase RAB-35 defines a third pathway that is required for the recognition and degradation of apoptotic cells
Abstract
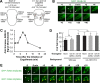
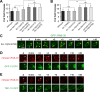
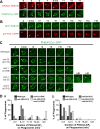
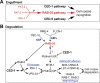
In metazoans, apoptotic cells are swiftly engulfed by phagocytes and degraded inside phagosomes. Multiple small GTPases in the Rab family are known to function in phagosome maturation by regulating vesicle trafficking. We discovered rab-35 as a new gene important for apoptotic cell clearance from a genetic screen targeting putative Rab GTPases in Caenorhabditis elegans. We further identified TBC-10 as a putative GTPase-activating protein (GAP), and FLCN-1 and RME-4 as two putative Guanine Nucleotide Exchange Factors (GEFs), for RAB-35. We found that RAB-35 was required for the efficient incorporation of early endosomes to phagosomes and for the timely degradation of apoptotic cell corpses. More specifically, RAB-35 promotes two essential events that initiate phagosome maturation: the switch of phagosomal membrane phosphatidylinositol species from PtdIns(4,5)P2 to PtdIns(3)P, and the recruitment of the small GTPase RAB-5 to phagosomal surfaces. These functions of RAB-35 were previously unknown. Remarkably, although the phagocytic receptor CED-1 regulates these same events, RAB-35 and CED-1 appear to function independently. Upstream of degradation, RAB-35 also facilitates the recognition of apoptotic cells independently of the known CED-1 and CED-5 pathways. RAB-35 localizes to extending pseudopods and is further enriched on nascent phagosomes, consistent with its dual roles in regulating apoptotic cell-recognition and phagosome maturation. Epistasis analyses indicate that rab-35 acts in parallel to both of the canonical ced-1/6/7 and ced-2/5/10/12 clearance pathways. We propose that RAB-35 acts as a robustness factor, defining a novel pathway that aids these canonical pathways in both the recognition and degradation of apoptotic cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







Comment in
-
C. elegans RAB-35: Dual roles in apoptotic cell clearance.PLoS Genet. 2018 Aug 23;14(8):e1007534. doi: 10.1371/journal.pgen.1007534. eCollection 2018 Aug. PLoS Genet. 2018. PMID: 30138327 Free PMC article. No abstract available.
References
-
- Lockshin RA, Williams CM. Programmed cell death—II. Endocrine potentiation of the breakdown of the intersegmental muscles of silkmoths. J Insect Physiol. 1964. August 1;10(4):643–9.
-
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983. November 1;100(1):64–119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

