Simultaneously Measuring Image Features and Resolution in Live-Cell STED Images
- PMID: 30139523
- PMCID: PMC6139878
- DOI: 10.1016/j.bpj.2018.07.028
Simultaneously Measuring Image Features and Resolution in Live-Cell STED Images
Abstract
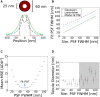
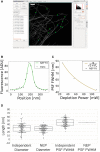
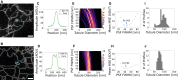
Reliable interpretation and quantification of cellular features in fluorescence microscopy requires an accurate estimate of microscope resolution. This is typically obtained by measuring the image of a nonbiological proxy for a point-like object, such as a fluorescent bead. Although appropriate for confocal microscopy, bead-based measurements are problematic for stimulated emission depletion microscopy and similar techniques where the resolution depends critically on the choice of fluorophore and acquisition parameters. In this article, we demonstrate that for a known geometry (e.g., tubules), the resolution can be measured in situ by fitting a model that accounts for both the point spread function (PSF) and the fluorophore distribution. To address the problem of coupling between tubule diameter and PSF width, we developed a technique called nested-loop ensemble PSF fitting. This approach enables extraction of the size of cellular features and the PSF width in fixed-cell and live-cell images without relying on beads or precalibration. Nested-loop ensemble PSF fitting accurately recapitulates microtubule diameter from stimulated emission depletion images and can measure the diameter of endoplasmic reticulum tubules in live COS-7 cells. Our algorithm has been implemented as a plugin for the PYthon Microscopy Environment, a freely available and open-source software.
Copyright © 2018 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Banterle N., Bui K.H., Beck M. Fourier ring correlation as a resolution criterion for super-resolution microscopy. J. Struct. Biol. 2013;183:363–367. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

