Comparative Genomics Approaches Accurately Predict Deleterious Variants in Plants
- PMID: 30139765
- PMCID: PMC6169392
- DOI: 10.1534/g3.118.200563
Comparative Genomics Approaches Accurately Predict Deleterious Variants in Plants
Abstract
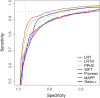
Recent advances in genome resequencing have led to increased interest in prediction of the functional consequences of genetic variants. Variants at phylogenetically conserved sites are of particular interest, because they are more likely than variants at phylogenetically variable sites to have deleterious effects on fitness and contribute to phenotypic variation. Numerous comparative genomic approaches have been developed to predict deleterious variants, but the approaches are nearly always assessed based on their ability to identify known disease-causing mutations in humans. Determining the accuracy of deleterious variant predictions in nonhuman species is important to understanding evolution, domestication, and potentially to improving crop quality and yield. To examine our ability to predict deleterious variants in plants we generated a curated database of 2,910 Arabidopsis thaliana mutants with known phenotypes. We evaluated seven approaches and found that while all performed well, their relative ranking differed from prior benchmarks in humans. We conclude that deleterious mutations can be reliably predicted in A. thaliana and likely other plant species, but that the relative performance of various approaches does not necessarily translate from one species to another.
Keywords: deleterious mutations; genome; phenotypes; training set.
Copyright © 2018 Kono et al.
Figures



Similar articles
-
The Role of Deleterious Substitutions in Crop Genomes.Mol Biol Evol. 2016 Sep;33(9):2307-17. doi: 10.1093/molbev/msw102. Epub 2016 Jun 14. Mol Biol Evol. 2016. PMID: 27301592 Free PMC article.
-
The Fate of Deleterious Variants in a Barley Genomic Prediction Population.Genetics. 2019 Dec;213(4):1531-1544. doi: 10.1534/genetics.119.302733. Epub 2019 Oct 25. Genetics. 2019. PMID: 31653677 Free PMC article.
-
Deleterious amino acid polymorphisms in Arabidopsis thaliana and rice.Theor Appl Genet. 2010 Jun;121(1):157-68. doi: 10.1007/s00122-010-1299-4. Epub 2010 Mar 3. Theor Appl Genet. 2010. PMID: 20198468
-
Genetic Costs of Domestication and Improvement.J Hered. 2018 Feb 14;109(2):103-116. doi: 10.1093/jhered/esx069. J Hered. 2018. PMID: 28992310 Review.
-
Evolutionary dynamics and adaptive benefits of deleterious mutations in crop gene pools.Trends Plant Sci. 2023 Jun;28(6):685-697. doi: 10.1016/j.tplants.2023.01.006. Epub 2023 Feb 8. Trends Plant Sci. 2023. PMID: 36764870 Review.
Cited by
-
Seed management using NGS technology to rapidly eliminate a deleterious allele from rice breeder seeds.Breed Sci. 2022 Dec;72(5):362-371. doi: 10.1270/jsbbs.22058. Epub 2022 Dec 13. Breed Sci. 2022. PMID: 36776441 Free PMC article.
-
PlaPPISite: a comprehensive resource for plant protein-protein interaction sites.BMC Plant Biol. 2020 Feb 6;20(1):61. doi: 10.1186/s12870-020-2254-4. BMC Plant Biol. 2020. PMID: 32028878 Free PMC article.
-
Biased Gene Conversion Constrains Adaptation in Arabidopsis thaliana.Genetics. 2020 Jul;215(3):831-846. doi: 10.1534/genetics.120.303335. Epub 2020 May 15. Genetics. 2020. PMID: 32414868 Free PMC article.
-
Multiple Sources of Introduction of North American Arabidopsis thaliana from across Eurasia.Mol Biol Evol. 2021 Dec 9;38(12):5328-5344. doi: 10.1093/molbev/msab268. Mol Biol Evol. 2021. PMID: 34499163 Free PMC article.
-
The patterns of deleterious mutations during the domestication of soybean.Nat Commun. 2021 Jan 4;12(1):97. doi: 10.1038/s41467-020-20337-3. Nat Commun. 2021. PMID: 33397978 Free PMC article.
References
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
