Chromatin de-condensation by switching substrate elasticity
- PMID: 30140058
- PMCID: PMC6107547
- DOI: 10.1038/s41598-018-31023-2
Chromatin de-condensation by switching substrate elasticity
Abstract
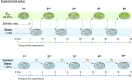
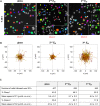
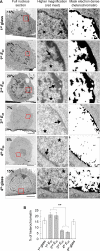
Mechanical properties of the cellular environment are known to influence cell fate. Chromatin de-condensation appears as an early event in cell reprogramming. Whereas the ratio of euchromatin versus heterochromatin can be increased chemically, we report herein for the first time that the ratio can also be increased by purely changing the mechanical properties of the microenvironment by successive 24 h-contact of the cells on a soft substrate alternated with relocation and growth for 7 days on a hard substrate. An initial contact with soft substrate caused massive SW480 cancer cell death by necrosis, whereas approximately 7% of the cells did survived exhibiting a high level of condensed chromatin (21% heterochromatin). However, four consecutive hard/soft cycles elicited a strong chromatin de-condensation (6% heterochromatin) correlating with an increase of cellular survival (approximately 90%). Furthermore, cell survival appeared to be reversible, indicative of an adaptive process rather than an irreversible gene mutation(s). This adaptation process is associated with modifications in gene expression patterns. A completely new approach for chromatin de-condensation, based only on mechanical properties of the microenvironment, without any drug mediation is presented.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

