Environment Controls LEE Regulation in Enteropathogenic Escherichia coli
- PMID: 30140259
- PMCID: PMC6094958
- DOI: 10.3389/fmicb.2018.01694
Environment Controls LEE Regulation in Enteropathogenic Escherichia coli
Abstract
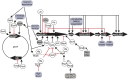
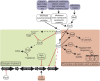
Enteropathogenic Escherichia coli (EPEC) is a significant cause of infant morbidity and mortality in developing regions of the world. Horizontally acquired genetic elements encode virulence structures, effectors, and regulators that promote bacterial colonization and disease. One such genetic element, the locus of enterocyte effacement (LEE), encodes the type three secretion system (T3SS) which acts as a bridge between bacterial and host cells to pass effector molecules that exert changes on the host. Due to its importance in EPEC virulence, regulation of the LEE has been of high priority and its investigation has elucidated many virulence regulators, including master regulator of the LEE Ler, H-NS, other nucleoid-associated proteins, GrlA, and PerC. Media type, environmental signals, sRNA signaling, metabolic processes, and stress responses have profound, strain-specific effects on regulators and LEE expression, and thus T3SS formation. Here we review virulence gene regulation in EPEC, which includes approaches for lessening disease by exploiting the elucidated regulatory pathways.
Keywords: EPEC; LEE; envelope stress; environment; metabolism; sRNA; virulence.
Figures
References
-
- Ahmed S. F., Shaheen H. I., Abdel-Messih I. A., Mostafa M., Putnam S. D., Kamal K. A., et al. (2014). The epidemiological and clinical characteristics of diarrhea associated with enteropathogenic, enteroaggregative and diffuse-adherent Escherichia coli in egyptian children. J. Trop. Pediatr. 60 397–400. 10.1093/tropej/fmu034 - DOI - PubMed
-
- Ashokkumar B., Kumar J. S., Hecht G. A., Said H. M. (2009). Enteropathogenic Escherichia coli inhibits intestinal vitamin B1 (thiamin) uptake: studies with human-derived intestinal epithelial Caco-2 cells. Am. J. Physiol. Gastrointest. Liver Physiol. 297 G825–G833. 10.1152/ajpgi.00250.2009 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

