Identification of expression quantitative trait loci associated with schizophrenia and affective disorders in normal brain tissue
- PMID: 30142156
- PMCID: PMC6126875
- DOI: 10.1371/journal.pgen.1007607
Identification of expression quantitative trait loci associated with schizophrenia and affective disorders in normal brain tissue
Abstract
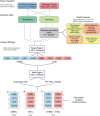
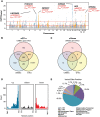
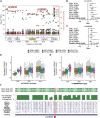
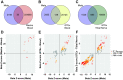
Schizophrenia and the affective disorders, here comprising bipolar disorder and major depressive disorder, are psychiatric illnesses that lead to significant morbidity and mortality worldwide. Whilst understanding of their pathobiology remains limited, large case-control studies have recently identified single nucleotide polymorphisms (SNPs) associated with these disorders. However, discerning the functional effects of these SNPs has been difficult as the associated causal genes are unknown. Here we evaluated whether schizophrenia and affective disorder associated-SNPs are correlated with gene expression within human brain tissue. Specifically, to identify expression quantitative trait loci (eQTLs), we leveraged disorder-associated SNPs identified from 11 genome-wide association studies with gene expression levels in post-mortem, neurologically-normal tissue from two independent human brain tissue expression datasets (UK Brain Expression Consortium (UKBEC) and Genotype-Tissue Expression (GTEx)). Utilizing stringent multi-region meta-analyses, we identified 2,224 cis-eQTLs associated with expression of 40 genes, including 11 non-coding RNAs. One cis-eQTL, rs16969968, results in a functionally disruptive missense mutation in CHRNA5, a schizophrenia-implicated gene. Importantly, comparing across tissues, we find that blood eQTLs capture < 10% of brain cis-eQTLs. Contrastingly, > 30% of brain-associated eQTLs are significant in tibial nerve. This study identifies putatively causal genes whose expression in region-specific tissue may contribute to the risk of schizophrenia and affective disorders.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- WHO. Mental Disorders: World Health Organization; 2014 [cited 2015 23 September]. http://www.who.int/mediacentre/factsheets/fs396/en/.
-
- Murray CJ, Vos T, Lozano R, Naghavi M, Flaxman AD, Michaud C, et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2197–223. 10.1016/S0140-6736(12)61689-4 . - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

