Challenges and approaches to predicting RNA with multiple functional structures
- PMID: 30143552
- PMCID: PMC6239171
- DOI: 10.1261/rna.067827.118
Challenges and approaches to predicting RNA with multiple functional structures
Abstract
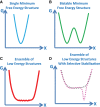
The revolution in sequencing technology demands new tools to interpret the genetic code. As in vivo transcriptome-wide chemical probing techniques advance, new challenges emerge in the RNA folding problem. The emphasis on one sequence folding into a single minimum free energy structure is fading as a new focus develops on generating RNA structural ensembles and identifying functional structural features in ensembles. This review describes an efficient combinatorially complete method and three free energy minimization approaches to predicting RNA structures with more than one functional fold, as well as two methods for analysis of a thermodynamics-based Boltzmann ensemble of structures. The review then highlights two examples of viral RNA 3'-UTR regions that fold into more than one conformation and have been characterized by single molecule fluorescence energy resonance transfer or NMR spectroscopy. These examples highlight the different approaches and challenges in predicting structure and function from sequence for RNA with multiple biological roles and folds. More well-defined examples and new metrics for measuring differences in RNA structures will guide future improvements in prediction of RNA structure and function from sequence.
Keywords: RNA conformational landscape; RNA folding; RNA free energy minimization; RNA structure prediction; in vivo genome-wide chemical probing.
© 2018 Schroeder; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures
References
-
- Backofen R, Gorodkin J, Hofacker IL, Stadler PF. 2018. Comparative RNA genomics. Methods Mol Biol 1704: 363–400. - PubMed
-
- Bailey S, Wichitwechkarn J, Johnson D, Reilly BE, Anderson DL, Bodley JW. 1990. Phylogenetic analysis and secondary structure of Bacillus subtilis bacteriophage RNA required for DNA packaging. J Biol Chem 265: 22365–22370. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
