Whole-genome sequencing and comparative analysis of two plant-associated strains of Rhodopseudomonas palustris (PS3 and YSC3)
- PMID: 30143697
- PMCID: PMC6109142
- DOI: 10.1038/s41598-018-31128-8
Whole-genome sequencing and comparative analysis of two plant-associated strains of Rhodopseudomonas palustris (PS3 and YSC3)
Abstract
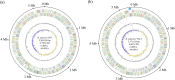
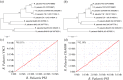
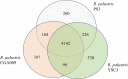
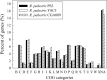
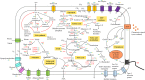
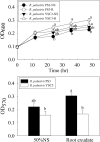
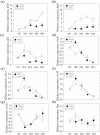
Rhodopseudomonas palustris strains PS3 and YSC3 are purple non-sulfur phototrophic bacteria isolated from Taiwanese paddy soils. PS3 has beneficial effects on plant growth and enhances the uptake efficiency of applied fertilizer nutrients. In contrast, YSC3 has no significant effect on plant growth. The genomic structures of PS3 and YSC3 are similar; each contains one circular chromosome that is 5,269,926 or 5,371,816 bp in size, with 4,799 or 4,907 protein-coding genes, respectively. In this study, a large class of genes involved in chemotaxis and motility was identified in both strains, and genes associated with plant growth promotion, such as nitrogen fixation-, IAA synthesis- and ACC deamination-associated genes, were also identified. We noticed that the growth rate, the amount of biofilm formation, and the relative expression levels of several chemotaxis-associated genes were significantly higher for PS3 than for YSC3 upon treatment with root exudates. These results indicate that PS3 responds better to the presence of plant hosts, which may contribute to the successful interactions of PS3 with plant hosts. Moreover, these findings indicate that the existence of gene clusters associated with plant growth promotion is required but not sufficient for a bacterium to exhibit phenotypes associated with plant growth promotion.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Kloepper, J. W. & Schroth, M. N. In Proceedings of the 4th international conference on plant pathogenic bacteria. (ed. Station de Pathologie Végétale et Phyto-Bactériologie) 879–882 (1978).
-
- Ahemad M, Kibret M. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J. King Saud Univ. Sci. 2014;26:1–20. doi: 10.1016/j.jksus.2013.05.001. - DOI
-
- Goswami D, Thakker JN, Dhandhukia PC. Portraying mechanics of plant growth promoting rhizobacteria (PGPR): a review. Cogent Food Agric. 2016;2:1127500–1127518.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

