Pathological missorting of endogenous MAPT/Tau in neurons caused by failure of protein degradation systems
- PMID: 30145931
- PMCID: PMC6984766
- DOI: 10.1080/15548627.2018.1509607
Pathological missorting of endogenous MAPT/Tau in neurons caused by failure of protein degradation systems
Abstract
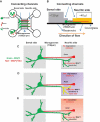
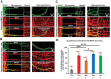
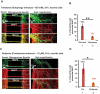
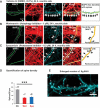
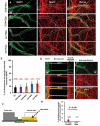
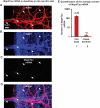
Missorting of MAPT/Tau represents one of the early signs of neurodegeneration in Alzheimer disease. The triggers for this are still a matter of debate. Here we investigated the sorting mechanisms of endogenous MAPT in mature primary neurons using microfluidic chambers (MFCs) where cell compartments can be observed separately. Blocking protein degradation pathways with proteasomal or autophagy inhibitors dramatically increased the missorting of MAPT in dendrites on the neuritic side, suggesting that degradation of MAPT in dendrites is a major determinant for the physiological axonal distribution of MAPT. Such missorted dendritic MAPT differed in its phosphorylation pattern from axonal MAPT. By contrast, enhancing autophagy or proteasomal pathways strongly reduced MAPT missorting, thereby confirming the role of protein degradation pathways in the polar distribution of MAPT. Dendritic missorting of MAPT by blocking protein degradation resulted in the loss of spines but not in overall cell toxicity. Inhibition of local protein synthesis in dendrites eliminated the missorting of MAPT, indicating that the accumulation of dendritic MAPT is locally generated. In support of this, a substantial fraction of Mapt/Tau mRNA was detected in dendrites. Taken together, our results indicate that the autophagy and proteasomal pathways play important roles in fine-tuning dendritic MAPT levels and thereby prevent synaptic toxicity caused by MAPT accumulation. Abbreviations Ani: anisomycin; Baf: bafilomycin A1; BSA: bovine serum albumin; cAMP: cyclic adenosine monophosphate; CHX: cycloheximide; DMSO: dimethyl sulfoxide; DIV: days in vitro; Epo: epoxomicin; E18: embryonic day 18; FISH: fluorescence in situ hybridization; IgG: immunoglobulin; kDa: kilodalton; Lac: lactacystin; LDH: lactate dehydrogenase; MFC: microfluidic chambers; MAPs: microtubule-associated proteins; MAPT/Tau: microtubule-associated protein tau; PVDF: polyvinylidene difluoride; PBS: phosphate-buffered saline; PRKA: protein kinase AMP-activated; RD150: round device 150; RT: room temperature; SDS: sodium dodecyl sulfate; SEM: standard error of the mean; Wor: wortmannin.
Keywords: Alzheimer disease; MAPT/Tau; autophagy; degradation; microfluidic chambers; proteasome.
Figures


References
-
- Braak E, Braak H, Mandelkow EM. A sequence of cytoskeleton changes related to the formation of neurofibrillary tangles and neuropil threads. Acta Neuropathol. 1994;87(6):554–567. PubMed PMID: 7522386. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
