Chromosome Segregation Fidelity in Epithelia Requires Tissue Architecture
- PMID: 30146160
- PMCID: PMC6151153
- DOI: 10.1016/j.cell.2018.07.042
Chromosome Segregation Fidelity in Epithelia Requires Tissue Architecture
Abstract
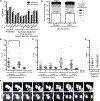
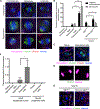
Much of our understanding of chromosome segregation is based on cell culture systems. Here, we examine the importance of the tissue environment for chromosome segregation by comparing chromosome segregation fidelity across several primary cell types in native and nonnative contexts. We discover that epithelial cells have increased chromosome missegregation outside of their native tissues. Using organoid culture systems, we show that tissue architecture, specifically integrin function, is required for accurate chromosome segregation. We find that tissue architecture enhances the correction of merotelic microtubule-kinetochore attachments, and this is especially important for maintaining chromosome stability in the polyploid liver. We propose that disruption of tissue architecture could underlie the widespread chromosome instability across epithelial cancers. Moreover, our findings highlight the extent to which extracellular context can influence intrinsic cellular processes and the limitations of cell culture systems for studying cells that naturally function within a tissue.
Keywords: aneuploidy; chromosome segregation; integrin; tissue architecture.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures


Comment in
-
Chromosome Segregation Fidelity Integrates Information across Scales.Dev Cell. 2018 Dec 3;47(5):525-526. doi: 10.1016/j.devcel.2018.11.027. Dev Cell. 2018. PMID: 30513291
References
-
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J-S, Yates JR III, Chan CSM, Drubin DG, and Barnes G (2002). Phospho-Regulation of Kinetochore-Microtubule Attachments by the Aurora Kinase Ipl1p. Cell 111, 163–172. - PubMed
-
- Cimini D, Cameron LA, and Salmon ED (2004). Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Current Biology 14, 2149–2155. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

