Accurate Recycling of Parental Histones Reproduces the Histone Modification Landscape during DNA Replication
- PMID: 30146316
- PMCID: PMC6202308
- DOI: 10.1016/j.molcel.2018.08.010
Accurate Recycling of Parental Histones Reproduces the Histone Modification Landscape during DNA Replication
Abstract
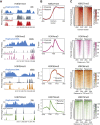
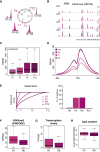
Chromatin organization is disrupted genome-wide during DNA replication. On newly synthesized DNA, nucleosomes are assembled from new naive histones and old modified histones. It remains unknown whether the landscape of histone post-translational modifications (PTMs) is faithfully copied during DNA replication or the epigenome is perturbed. Here we develop chromatin occupancy after replication (ChOR-seq) to determine histone PTM occupancy immediately after DNA replication and across the cell cycle. We show that H3K4me3, H3K36me3, H3K79me3, and H3K27me3 positional information is reproduced with high accuracy on newly synthesized DNA through histone recycling. Quantitative ChOR-seq reveals that de novo methylation to restore H3K4me3 and H3K27me3 levels occurs across the cell cycle with mark- and locus-specific kinetics. Collectively, this demonstrates that accurate parental histone recycling preserves positional information and allows PTM transmission to daughter cells while modification of new histones gives rise to complex epigenome fluctuations across the cell cycle that could underlie cell-to-cell heterogeneity.
Keywords: ChOR-seq; H3K27me3; H3K4me3; cell cycle; chromatin replication; epigenetics; epigenome maintenance; histone modification; histone recycling; quantitative ChIP-seq.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Alabert C., Groth A. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 2012;13:153–167. - PubMed
-
- Alabert C., Bukowski-Wills J.C., Lee S.B., Kustatscher G., Nakamura K., de Lima Alves F., Menard P., Mejlvang J., Rappsilber J., Groth A. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 2014;16:281–293. - PMC - PubMed
-
- Allis C.D., Jenuwein T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016;17:487–500. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

