The miR 495-UBE2C-ABCG2/ERCC1 axis reverses cisplatin resistance by downregulating drug resistance genes in cisplatin-resistant non-small cell lung cancer cells
- PMID: 30146342
- PMCID: PMC6419862
- DOI: 10.1016/j.ebiom.2018.08.001
The miR 495-UBE2C-ABCG2/ERCC1 axis reverses cisplatin resistance by downregulating drug resistance genes in cisplatin-resistant non-small cell lung cancer cells
Retraction in
-
Retraction notice to 'The miR 495-UBE2C-ABCG2/ERCC1 axis reverses cisplatin resistance by downregulating drug resistance genes in cisplatin-resistant non-small cell lung cancer cells' [EBioMedicine 35 (2018) 204-221].EBioMedicine. 2021 Jan;63:103168. doi: 10.1016/j.ebiom.2020.103168. EBioMedicine. 2021. PMID: 33487222 Free PMC article. No abstract available.
Abstract
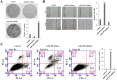
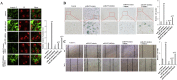
Cisplatin (DDP) resistance has become the leading cause of mortality in non-small cell lung cancer (NSCLC). miRNA dysregulation significantly contributes to tumor progression. In this study, we found that miR-495 was significantly downregulated in lung cancer tissue specimens. This study aimed to elucidate the functions, direct target genes, and molecular mechanisms of miR-495 in lung cancer. miR-495 downregulated its substrate UBE2C through direct interaction with UBE2C 3'- untranslated region. UBE2C is a proto-oncogene activated in lung cancer; however, its role in chemotherapeutic resistance is unclear. Herein, UBE2C expression levels were higher in DDP-resistant NSCLC cells; this was associated with the proliferation, invasion, and DDP resistance in induced cisplatin-resistant NSCLC cells. Furthermore, epithelial-mesenchymal transitions (EMT) contributed to DDP resistance. Moreover, UBE2C knockdown downregulated vimentin. In contrast, E-cadherin was upregulated. Importantly, miR-495 and UBE2C were associated with cisplatin resistance. We attempted to evaluate their effects on cell proliferation and cisplatin resistance. We also performed EMT, cell migration, and invasion assays in DDP-resistant NSCLC cells overexpressing miR-495 and under-expressing UBE2C. Furthermore, in silico assays coupled with western blotting and luciferase assays revealed that UBE2C directly binds to the 5'-UTR of the drug-resistance genes ABCG2 and ERCC1. Furthermore, miR-495 downregulated ABCG2 and ERCC1 via regulation of UBE2C. Together, the present results indicate that the miR495-UBE2C-ABCG2/ERCC1 axis reverses DDP resistance via downregulation of anti-drug genes and reducing EMT in DDP-resistant NSCLC cells.
Keywords: ABCG2; Cisplatin resistant; EMT; ERCC1; MicroRNA-495; UBE2C.
Copyright © 2018 The Author(s). Published by Elsevier B.V. All rights reserved.
Figures













References
-
- Barton M.K. Human immunodeficiency virus status has no effect on survival in patients with non-small cell lung cancer. CA Cancer J Clin. 2013;63(3):145–146. - PubMed
-
- Barton M.K. Adjuvant chemotherapy benefits older and younger non-small cell lung cancer patients alike. CA Cancer J Clin. 2012;62(5):279–280. - PubMed
-
- Ramalingam S.S., Owonikoko T.K., Khuri F.R. Lung cancer: new biological insights and recent therapeutic advances. CA Cancer J Clin. 2011;61(2):91–112. - PubMed
-
- McCarthy N. MicroRNA: lacking in maturity. Nat Rev Cancer. 2013;13(6):377. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

