A Comprehensive Roadmap of Murine Spermatogenesis Defined by Single-Cell RNA-Seq
- PMID: 30146481
- PMCID: PMC6713459
- DOI: 10.1016/j.devcel.2018.07.025
A Comprehensive Roadmap of Murine Spermatogenesis Defined by Single-Cell RNA-Seq
Abstract
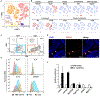
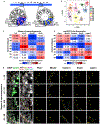
Spermatogenesis requires intricate interactions between the germline and somatic cells. Within a given cross section of a seminiferous tubule, multiple germ and somatic cell types co-occur. This cellular heterogeneity has made it difficult to profile distinct cell types at different stages of development. To address this challenge, we collected single-cell RNA sequencing data from ∼35,000 cells from the adult mouse testis and identified all known germ and somatic cells, as well as two unexpected somatic cell types. Our analysis revealed a continuous developmental trajectory of germ cells from spermatogonia to spermatids and identified candidate transcriptional regulators at several transition points during differentiation. Focused analyses delineated four subtypes of spermatogonia and nine subtypes of Sertoli cells; the latter linked to histologically defined developmental stages over the seminiferous epithelial cycle. Overall, this high-resolution cellular atlas represents a community resource and foundation of knowledge to study germ cell development and in vivo gametogenesis.
Keywords: germ cell developmental trajectory; germ cell differentiation; heterogeneity; single-cell RNA-seq; spermatogenesis; spermatogonial stem cell; testis niche.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests
The authors have no competing interests.
Figures





References
-
- Anderson EL, Baltus AE, Roepers-Gajadien HL, Hassold TJ, de Rooij DG, van Pelt AM, and Page DC (2008). Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proceedings of the National Academy of Sciences of the United States of America 105, 14976–14980. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

