Chemical Screening Identifies Enhancers of Mutant Oligodendrocyte Survival and Unmasks a Distinct Pathological Phase in Pelizaeus-Merzbacher Disease
- PMID: 30146490
- PMCID: PMC6135742
- DOI: 10.1016/j.stemcr.2018.07.015
Chemical Screening Identifies Enhancers of Mutant Oligodendrocyte Survival and Unmasks a Distinct Pathological Phase in Pelizaeus-Merzbacher Disease
Abstract
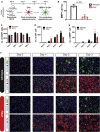
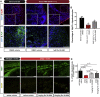
Pelizaeus-Merzbacher disease (PMD) is a fatal X-linked disorder caused by loss of myelinating oligodendrocytes and consequent hypomyelination. The underlying cellular and molecular dysfunctions are not fully defined, but therapeutic enhancement of oligodendrocyte survival could restore functional myelination in patients. Here we generated pure, scalable quantities of induced pluripotent stem cell-derived oligodendrocyte progenitor cells (OPCs) from a severe mouse model of PMD, Plp1jimpy. Temporal phenotypic and transcriptomic studies defined an early pathological window characterized by endoplasmic reticulum (ER) stress and cell death as OPCs exit their progenitor state. High-throughput phenotypic screening identified a compound, Ro 25-6981, which modulates the ER stress response and rescues mutant oligodendrocyte survival in jimpy, in vitro and in vivo, and in human PMD oligocortical spheroids. Surprisingly, increasing oligodendrocyte survival did not restore subsequent myelination, revealing a second pathological phase. Collectively, our work shows that PMD oligodendrocyte loss can be rescued pharmacologically and defines a need for multifactorial intervention to restore myelination.
Keywords: PLP1; Pelizaeus-Merzbacher disease; endoplasmic reticulum stress; high-throughput screening; iPSC disease modeling; myelin; oligodendrocyte progenitor cells; oligodendrocytes; proteolipid protein 1; rare disease.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





Similar articles
-
Modeling the Mutational and Phenotypic Landscapes of Pelizaeus-Merzbacher Disease with Human iPSC-Derived Oligodendrocytes.Am J Hum Genet. 2017 Apr 6;100(4):617-634. doi: 10.1016/j.ajhg.2017.03.005. Epub 2017 Mar 30. Am J Hum Genet. 2017. PMID: 28366443 Free PMC article.
-
Involvement of ER stress in dysmyelination of Pelizaeus-Merzbacher Disease with PLP1 missense mutations shown by iPSC-derived oligodendrocytes.Stem Cell Reports. 2014 Apr 24;2(5):648-61. doi: 10.1016/j.stemcr.2014.03.007. eCollection 2014 May 6. Stem Cell Reports. 2014. PMID: 24936452 Free PMC article.
-
Oligodendrocyte Death in Pelizaeus-Merzbacher Disease Is Rescued by Iron Chelation.Cell Stem Cell. 2019 Oct 3;25(4):531-541.e6. doi: 10.1016/j.stem.2019.09.003. Cell Stem Cell. 2019. PMID: 31585094 Free PMC article.
-
Neurogenetics of Pelizaeus-Merzbacher disease.Handb Clin Neurol. 2018;148:701-722. doi: 10.1016/B978-0-444-64076-5.00045-4. Handb Clin Neurol. 2018. PMID: 29478609 Free PMC article. Review.
-
Pelizaeus-Merzbacher disease: on the cusp of myelin medicine.Trends Mol Med. 2024 May;30(5):459-470. doi: 10.1016/j.molmed.2024.03.005. Epub 2024 Apr 5. Trends Mol Med. 2024. PMID: 38582621 Free PMC article. Review.
Cited by
-
Unfolded protein response in myelin disorders.Neural Regen Res. 2020 Apr;15(4):636-645. doi: 10.4103/1673-5374.266903. Neural Regen Res. 2020. PMID: 31638085 Free PMC article. Review.
-
Enhancers of Human and Rodent Oligodendrocyte Formation Predominantly Induce Cholesterol Precursor Accumulation.ACS Chem Biol. 2022 Aug 19;17(8):2188-2200. doi: 10.1021/acschembio.2c00330. Epub 2022 Jul 14. ACS Chem Biol. 2022. PMID: 35833657 Free PMC article.
-
Modeling vanishing white matter disease with patient-derived induced pluripotent stem cells reveals astrocytic dysfunction.CNS Neurosci Ther. 2019 Jun;25(6):759-771. doi: 10.1111/cns.13107. Epub 2019 Feb 5. CNS Neurosci Ther. 2019. PMID: 30720246 Free PMC article.
-
Alterations in oligodendrocyte transcriptional networks reveal region-specific vulnerabilities to neurological disease.iScience. 2023 Mar 8;26(4):106358. doi: 10.1016/j.isci.2023.106358. eCollection 2023 Apr 21. iScience. 2023. PMID: 36994077 Free PMC article.
-
High-throughput screening for myelination promoting compounds using human stem cell-derived oligodendrocyte progenitor cells.iScience. 2023 Feb 8;26(3):106156. doi: 10.1016/j.isci.2023.106156. eCollection 2023 Mar 17. iScience. 2023. PMID: 36852281 Free PMC article.
References
-
- Baron W., Hoekstra D. On the biogenesis of myelin membranes: sorting, trafficking and cell polarity. FEBS Lett. 2010;584:1760–1770. - PubMed
-
- Baumann N., Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol. Rev. 2001;81:871–927. - PubMed
-
- Boyce M., Bryant K.F., Jousse C., Long K., Harding H.P., Scheuner D., Kaufman R.J., Ma D., Coen D.M., Ron D. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

