Salmonella Typhimurium Invalidated for the Three Currently Known Invasion Factors Keeps Its Ability to Invade Several Cell Models
- PMID: 30148118
- PMCID: PMC6095967
- DOI: 10.3389/fcimb.2018.00273
Salmonella Typhimurium Invalidated for the Three Currently Known Invasion Factors Keeps Its Ability to Invade Several Cell Models
Abstract
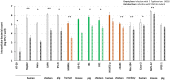
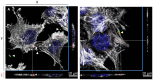
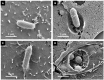
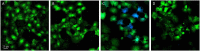
To establish an infection, Salmonella has to interact with eukaryotic cells. Invasion of non-phagocytic cells (i.e., epithelial, fibroblast and endothelial cells) involves either a trigger or a zipper mechanism mediated by the T3SS-1 or the invasin Rck, respectively. Another outer membrane protein, PagN, was also implicated in the invasion. However, other unknown invasion factors have been previously suggested. Our goal was to evaluate the invasion capability of a Salmonella Typhimurium strain invalidated for the three known invasion factors. Non-phagocytic cell lines of several animal origins were tested in a gentamicin protection assay. In most cells, we observed a drastic decrease in the invasion rate between the wild-type and the triple mutant. However, in five cell lines, the triple mutant invaded cells at a similarly high level to the wild-type, suggesting the existence of unidentified invasion factors. For the wild-type and the triple mutant, scanning-electron microscopy, confocal imaging and use of biochemical inhibitors confirmed their cellular uptake and showed a zipper-like mechanism of internalization involving both clathrin- and non-clathrin-dependent pathways. Despite a functional T3SS-1, the wild-type bacteria seemed to use the same entry route as the mutant in our cell model. All together, these results demonstrate the existence of unknown Salmonella invasion factors, which require further characterization.
Keywords: T3SS-1; T3SS-1 independent invasion; cell models; entry mechanism; trigger; zipper.
Figures


Similar articles
-
Investigation of the invasion mechanism mediated by the outer membrane protein PagN of Salmonella Typhimurium.BMC Microbiol. 2021 May 21;21(1):153. doi: 10.1186/s12866-021-02187-1. BMC Microbiol. 2021. PMID: 34020586 Free PMC article.
-
A large panel of chicken cells are invaded in vivo by Salmonella Typhimurium even when depleted of all known invasion factors.Open Biol. 2021 Nov;11(11):210117. doi: 10.1098/rsob.210117. Epub 2021 Nov 17. Open Biol. 2021. PMID: 34784793 Free PMC article.
-
Heterogeneity of type III secretion system (T3SS)-1-independent entry mechanisms used by Salmonella Enteritidis to invade different cell types.Microbiology (Reading). 2011 Mar;157(Pt 3):839-847. doi: 10.1099/mic.0.044941-0. Epub 2010 Nov 25. Microbiology (Reading). 2011. PMID: 21109565
-
An Updated View on the Rck Invasin of Salmonella: Still Much to Discover.Front Cell Infect Microbiol. 2017 Dec 8;7:500. doi: 10.3389/fcimb.2017.00500. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 29276700 Free PMC article. Review.
-
Virulence Factors in Salmonella Typhimurium: The Sagacity of a Bacterium.Curr Microbiol. 2019 Jun;76(6):762-773. doi: 10.1007/s00284-018-1510-4. Epub 2018 May 21. Curr Microbiol. 2019. PMID: 29785632 Review.
Cited by
-
Intracellular Survival and Pathogenicity Modulation of Salmonella Lon, CpxR, and RfaL Mutants Used as Live Bacterial Vectors under Abiotic Stress, Unveiling the Link between Stress Response and Virulence in Epithelial Cells.Int J Mol Sci. 2024 Aug 21;25(16):9056. doi: 10.3390/ijms25169056. Int J Mol Sci. 2024. PMID: 39201742 Free PMC article.
-
Chitinase A, a tightly regulated virulence factor of Salmonella enterica serovar Typhimurium, is actively secreted by a Type 10 Secretion System.PLoS Pathog. 2023 Apr 5;19(4):e1011306. doi: 10.1371/journal.ppat.1011306. eCollection 2023 Apr. PLoS Pathog. 2023. PMID: 37018381 Free PMC article.
-
Human tetraspanin CD81 facilitates invasion of Salmonella enterica into human epithelial cells.Virulence. 2024 Dec;15(1):2399792. doi: 10.1080/21505594.2024.2399792. Epub 2024 Sep 24. Virulence. 2024. PMID: 39239914 Free PMC article.
-
Cross-Talk Between the Intestinal Epithelium and Salmonella Typhimurium.Front Microbiol. 2022 Jun 6;13:906238. doi: 10.3389/fmicb.2022.906238. eCollection 2022. Front Microbiol. 2022. PMID: 35733975 Free PMC article. Review.
-
Activity of a Bacteriophage Cocktail to Control Salmonella Growth Ex Vivo in Avian, Porcine, and Human Epithelial Cell Cultures.Phage (New Rochelle). 2023 Mar 1;4(1):11-25. doi: 10.1089/phage.2023.0001. Epub 2023 Mar 17. Phage (New Rochelle). 2023. PMID: 37214653 Free PMC article.
References
-
- Charpentier X., Oswald E. (2004). Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J. Bacteriol. 186, 5486–5495. 10.1128/JB.186.16.5486-5495.2004 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

