Chemoproteomic identification of molecular targets of antifungal prototypes, thiosemicarbazide and a camphene derivative of thiosemicarbazide, in Paracoccidioides brasiliensis
- PMID: 30148835
- PMCID: PMC6110461
- DOI: 10.1371/journal.pone.0201948
Chemoproteomic identification of molecular targets of antifungal prototypes, thiosemicarbazide and a camphene derivative of thiosemicarbazide, in Paracoccidioides brasiliensis
Abstract
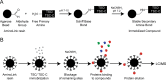
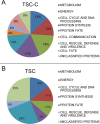
Paracoccidioidomycosis (PCM) is a neglected human systemic disease caused by species of the genus Paracoccidioides. The disease attacks the host's lungs and may disseminate to many other organs. Treatment involves amphotericin B, sulfadiazine, trimethoprim-sulfamethoxazole, itraconazole, ketoconazole, or fluconazole. The treatment duration is usually long, from 6 months to 2 years, and many adverse effects may occur in relation to the treatment; co-morbidities and poor treatment adherence have been noted. Therefore, the discovery of more effective and less toxic drugs is needed. Thiosemicarbazide (TSC) and a camphene derivative of thiosemicarbazide (TSC-C) were able to inhibit P. brasiliensis growth at a low dosage and were not toxic to fibroblast cells. In order to investigate the mode of action of those compounds, we used a chemoproteomic approach to determine which fungal proteins were bound to each of these compounds. The compounds were able to inhibit the activities of the enzyme formamidase and interfered in P. brasiliensis dimorphism. In comparison with the transcriptomic and proteomic data previously obtained by our group, we determined that TSC and TSC-C were multitarget compounds that exerted effects on the electron-transport chain and cell cycle regulation, increased ROS formation, inhibited proteasomes and peptidases, modulated glycolysis, lipid, protein and carbohydrate metabolisms, and caused suppressed the mycelium to yeast transition.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

