Errors in protein synthesis increase the level of saturated fatty acids and affect the overall lipid profiles of yeast
- PMID: 30148852
- PMCID: PMC6110467
- DOI: 10.1371/journal.pone.0202402
Errors in protein synthesis increase the level of saturated fatty acids and affect the overall lipid profiles of yeast
Abstract
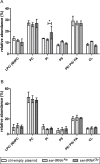
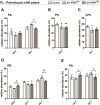
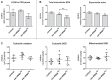
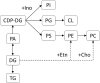
The occurrence of protein synthesis errors (mistranslation) above the typical mean mistranslation level of 10-4 is mostly deleterious to yeast, zebrafish and mammal cells. Previous yeast studies have shown that mistranslation affects fitness and deregulates genes related to lipid metabolism, but there is no experimental proof that such errors alter yeast lipid profiles. We engineered yeast strains to misincorporate serine at alanine and glycine sites on a global scale and evaluated the putative effects on the lipidome. Lipids from whole cells were extracted and analysed by thin layer chromatography (TLC), liquid chromatography-mass spectrometry(LC-MS) and gas chromatography (GC). Oxidative damage, fatty acid desaturation and membrane fluidity changes were screened to identify putative alterations in lipid profiles in both logarithmic (fermentative) and post-diauxic shift (respiratory) phases. There were alterations in several lipid classes, namely lyso-phosphatidylcholine, phosphatidic acid, phosphatidylethanolamine, phosphatidylinositol, phosphatidylserine, and triglyceride, and in the fatty acid profiles, namely C16:1, C16:0, C18:1 and C18:0. Overall, the relative content of lipid species with saturated FA increased in detriment of those with unsaturated fatty acids. The expression of the OLE1 mRNA was deregulated, but phospholipid fluidity changes were not observed. These data expand current knowledge of mistranslation biology and highlight its putative roles in human diseases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

