Characterization of Unknown Orthobunya-Like Viruses from India
- PMID: 30149496
- PMCID: PMC6165560
- DOI: 10.3390/v10090451
Characterization of Unknown Orthobunya-Like Viruses from India
Abstract
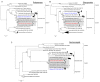
Next-generation sequencing (NGS) of agents causing idiopathic human diseases has been crucial in the identification of novel viruses. This study describes the isolation and characterization of two novel orthobunyaviruses obtained from a jungle myna and a paddy bird from Karnataka State, India. Using an NGS approach, these isolates were classified as Cat Que and Balagodu viruses belonging to the Manzanilla clade of the Simbu serogroup. Closely related viruses in the Manzanilla clade have been isolated from mosquitos, humans, birds, and pigs across a wide geographic region. Since Orthobunyaviruses exhibit high reassortment frequency and can cause acute, self-limiting febrile illness, these data suggest that human and livestock infections of the Oya/Cat Que/Manzanilla virus may be more widespread and/or under-reported than anticipated. It therefore becomes imperative to identify novel and unknown viruses in order to understand their role in human and animal pathogenesis. The current study is a step forward in this regard and would act as a prototype method for isolation, identification and detection of several other emerging viruses.
Keywords: Cat Que virus; Manzanilla virus; Orthobunyavirus; pathogen discovery.
Conflict of interest statement
The authors declare no competing conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Figures



References
-
- Piantadosi A., Mukerji S.S., Chitneni P., Cho T.A., Cosimi L.A., Hung D.T., Goldberg M.B., Sabeti P.C., Kuritzkes D.R., Grad Y.H. Metagenomic Sequencing of an Echovirus 30 Genome From Cerebrospinal Fluid of a Patient With Aseptic Meningitis and Orchitis. Open Forum. Infect. Dis. 2017;4:ofx138. doi: 10.1093/ofid/ofx138. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

