Three Distinct Contact-Dependent Growth Inhibition Systems Mediate Interbacterial Competition by the Cystic Fibrosis Pathogen Burkholderia dolosa
- PMID: 30150233
- PMCID: PMC6199481
- DOI: 10.1128/JB.00428-18
Three Distinct Contact-Dependent Growth Inhibition Systems Mediate Interbacterial Competition by the Cystic Fibrosis Pathogen Burkholderia dolosa
Abstract
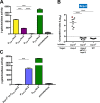
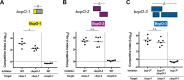
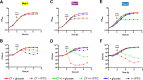
The respiratory tracts of individuals afflicted with cystic fibrosis (CF) harbor complex polymicrobial communities. By an unknown mechanism, species of the Gram-negative Burkholderia cepacia complex, such as Burkholderia dolosa, can displace other bacteria in the CF lung, causing cepacia syndrome, which has a poor prognosis. The genome of Bdolosa strain AU0158 (BdAU0158) contains three loci that are predicted to encode contact-dependent growth inhibition (CDI) systems. CDI systems function by translocating the toxic C terminus of a large exoprotein directly into target cells, resulting in growth inhibition or death unless the target cells produce a cognate immunity protein. We demonstrate here that each of the three bcpAIOB loci in BdAU0158 encodes a distinct CDI system that mediates interbacterial competition in an allele-specific manner. While only two of the three bcpAIOB loci were expressed under the in vitro conditions tested, the third conferred immunity under these conditions due to the presence of an internal promoter driving expression of the bcpI gene. One BdAU0158 bcpAIOB allele is highly similar to bcpAIOB in Burkholderia thailandensis strain E264 (BtE264), and we showed that their BcpI proteins are functionally interchangeable, but contact-dependent signaling (CDS) phenotypes were not observed in BdAU0158. Our findings suggest that the CDI systems of BdAU0158 may provide this pathogen an ecological advantage during polymicrobial infections of the CF respiratory tract.IMPORTANCE Human-associated polymicrobial communities can promote health and disease, and interbacterial interactions influence the microbial ecology of such communities. Polymicrobial infections of the cystic fibrosis respiratory tract impair lung function and lead to the death of individuals suffering from this disorder; therefore, a greater understanding of these microbial communities is necessary for improving treatment strategies. Bacteria utilize contact-dependent growth inhibition systems to kill neighboring competitors and maintain their niche within multicellular communities. Several cystic fibrosis pathogens have the potential to gain an ecological advantage during infection via contact-dependent growth inhibition systems, including Burkholderia dolosa Our research is significant, as it has identified three functional contact-dependent growth inhibition systems in Bdolosa that may provide this pathogen a competitive advantage during polymicrobial infections.
Keywords: Bcc; Burkholderia; contact-dependent inhibition; interbacterial competition; two-partner secretion.
Copyright © 2018 American Society for Microbiology.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

