Transcriptional switch for programmed cell death in pith parenchyma of sorghum stems
- PMID: 30150370
- PMCID: PMC6140496
- DOI: 10.1073/pnas.1807501115
Transcriptional switch for programmed cell death in pith parenchyma of sorghum stems
Abstract
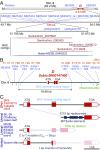
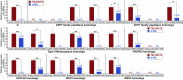
Pith parenchyma cells store water in various plant organs. These cells are especially important for producing sugar and ethanol from the sugar juice of grass stems. In many plants, the death of pith parenchyma cells reduces their stem water content. Previous studies proposed that a hypothetical D gene might be responsible for the death of stem pith parenchyma cells in Sorghum bicolor, a promising energy grass, although its identity and molecular function are unknown. Here, we identify the D gene and note that it is located on chromosome 6 in agreement with previous predictions. Sorghum varieties with a functional D allele had stems enriched with dry, dead pith parenchyma cells, whereas those with each of six independent nonfunctional D alleles had stems enriched with juicy, living pith parenchyma cells. D expression was spatiotemporally coupled with the appearance of dead, air-filled pith parenchyma cells in sorghum stems. Among D homologs that are present in flowering plants, Arabidopsis ANAC074 also is required for the death of stem pith parenchyma cells. D and ANAC074 encode previously uncharacterized NAC transcription factors and are sufficient to ectopically induce programmed death of Arabidopsis culture cells via the activation of autolytic enzymes. Taken together, these results indicate that D and its Arabidopsis ortholog, ANAC074, are master transcriptional switches that induce programmed death of stem pith parenchyma cells. Thus, targeting the D gene will provide an approach to breeding crops for sugar and ethanol production.
Keywords: NAC transcription factor; pith parenchyma; programmed cell death; sorghum; stem.
Conflict of interest statement
Conflict of interest statement: J.-i. Yonemaru, H.K., T.M., N.T., and S.K. are listed on a patent based on this work (Japanese patent P2015-025241), which was filed by NARO.
Figures





References
-
- Beers EP. Programmed cell death during plant growth and development. Cell Death Differ. 1997;4:649–661. - PubMed
-
- Carr SM, Jaffe MJ. Autolysis in herbaceous, dicotyledonous plants: Experimental manipulation of pith autolysis in several cultivated species. Ann Bot. 1995;75:587–592.
-
- Slewinski TL. Non-structural carbohydrate partitioning in grass stems: A target to increase yield stability, stress tolerance, and biofuel production. J Exp Bot. 2012;63:4647–4670. - PubMed
-
- Aloni B, Pressman E. Stem pithiness in tomato plants: The effect of water-stress and the role of abscisic-acid. Physiol Plant. 1981;51:39–44.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

