BRAF/MAPK and GSK3 signaling converges to control MITF nuclear export
- PMID: 30150413
- PMCID: PMC6140509
- DOI: 10.1073/pnas.1810498115
BRAF/MAPK and GSK3 signaling converges to control MITF nuclear export
Abstract
The close integration of the MAPK, PI3K, and WNT signaling pathways underpins much of development and is deregulated in cancer. In principle, combinatorial posttranslational modification of key lineage-specific transcription factors would be an effective means to integrate critical signaling events. Understanding how this might be achieved is central to deciphering the impact of microenvironmental cues in development and disease. The microphthalmia-associated transcription factor MITF plays a crucial role in the development of melanocytes, the retinal pigment epithelium, osteoclasts, and mast cells and acts as a lineage survival oncogene in melanoma. MITF coordinates survival, differentiation, cell-cycle progression, cell migration, metabolism, and lysosome biogenesis. However, how the activity of this key transcription factor is controlled remains poorly understood. Here, we show that GSK3, downstream from both the PI3K and Wnt pathways, and BRAF/MAPK signaling converges to control MITF nuclear export. Phosphorylation of the melanocyte MITF-M isoform in response to BRAF/MAPK signaling primes for phosphorylation by GSK3, a kinase inhibited by both PI3K and Wnt signaling. Dual phosphorylation, but not monophosphorylation, then promotes MITF nuclear export by activating a previously unrecognized hydrophobic export signal. Nonmelanocyte MITF isoforms exhibit poor regulation by MAPK signaling, but instead their export is controlled by mTOR. We uncover here an unanticipated mode of MITF regulation that integrates the output of key developmental and cancer-associated signaling pathways to gate MITF flux through the import-export cycle. The results have significant implications for our understanding of melanoma progression and stem cell renewal.
Keywords: GSK3; MAPK; MITF; melanoma; nuclear export.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
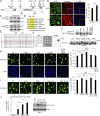
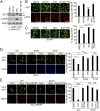
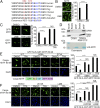
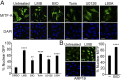
Comment in
-
Microphthalmia-associated transcription factor phosphorylation: Cross talk between GSK3 and MAPK signaling.Pigment Cell Melanoma Res. 2019 May;32(3):345-347. doi: 10.1111/pcmr.12765. Epub 2019 Jan 24. Pigment Cell Melanoma Res. 2019. PMID: 30633855 No abstract available.
References
-
- Garraway LA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. - PubMed
-
- Hodgkinson CA, et al. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. - PubMed
-
- Carreira S, et al. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 2005;433:764–769. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- C38302/A12981/CRUK_/Cancer Research UK/United Kingdom
- P01 CA128814/CA/NCI NIH HHS/United States
- MC_PC_U127585840/MRC_/Medical Research Council/United Kingdom
- R01 CA098571/CA/NCI NIH HHS/United States
- 19200/VAC_/Versus Arthritis/United Kingdom
- MC_U127585840/MRC_/Medical Research Council/United Kingdom
- MR/N010051/1/MRC_/Medical Research Council/United Kingdom
- 095751/Z/11/Z/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- MC_UU_00007/9/MRC_/Medical Research Council/United Kingdom
- R01 CA080728/CA/NCI NIH HHS/United States
- MP/19200/ARC_/Arthritis Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

