Jasmonates: News on Occurrence, Biosynthesis, Metabolism and Action of an Ancient Group of Signaling Compounds
- PMID: 30150593
- PMCID: PMC6164985
- DOI: 10.3390/ijms19092539
Jasmonates: News on Occurrence, Biosynthesis, Metabolism and Action of an Ancient Group of Signaling Compounds
Abstract
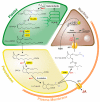
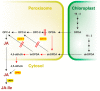
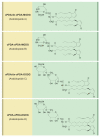
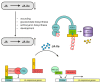
: Jasmonic acid (JA) and its related derivatives are ubiquitously occurring compounds of land plants acting in numerous stress responses and development. Recent studies on evolution of JA and other oxylipins indicated conserved biosynthesis. JA formation is initiated by oxygenation of α-linolenic acid (α-LeA, 18:3) or 16:3 fatty acid of chloroplast membranes leading to 12-oxo-phytodienoic acid (OPDA) as intermediate compound, but in Marchantiapolymorpha and Physcomitrellapatens, OPDA and some of its derivatives are final products active in a conserved signaling pathway. JA formation and its metabolic conversion take place in chloroplasts, peroxisomes and cytosol, respectively. Metabolites of JA are formed in 12 different pathways leading to active, inactive and partially active compounds. The isoleucine conjugate of JA (JA-Ile) is the ligand of the receptor component COI1 in vascular plants, whereas in the bryophyte M. polymorpha COI1 perceives an OPDA derivative indicating its functionally conserved activity. JA-induced gene expressions in the numerous biotic and abiotic stress responses and development are initiated in a well-studied complex regulation by homeostasis of transcription factors functioning as repressors and activators.
Keywords: JA biosynthetic enzymes; JA bypass; JA signaling; Jasmonic acid (JA) metabolites; active JA compounds; occurrence; transcription factors.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Wasternack C. How jasmonates earned their laurels: Past and present. J. Plant Growth Regul. 2015;34:761–794. doi: 10.1007/s00344-015-9526-5. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

