Revealing the mechanisms of membrane protein export by virulence-associated bacterial secretion systems
- PMID: 30150748
- PMCID: PMC6110835
- DOI: 10.1038/s41467-018-05969-w
Revealing the mechanisms of membrane protein export by virulence-associated bacterial secretion systems
Abstract
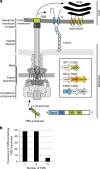
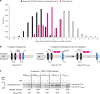
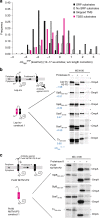
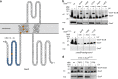
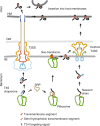
Many bacteria export effector proteins fulfilling their function in membranes of a eukaryotic host. These effector membrane proteins appear to contain signals for two incompatible bacterial secretion pathways in the same protein: a specific export signal, as well as transmembrane segments that one would expect to mediate targeting to the bacterial inner membrane. Here, we show that the transmembrane segments of effector proteins of type III and type IV secretion systems indeed integrate in the membrane as required in the eukaryotic host, but that their hydrophobicity in most instances is just below the threshold required for mediating targeting to the bacterial inner membrane. Furthermore, we show that binding of type III secretion chaperones to both the effector's chaperone-binding domain and adjacent hydrophobic transmembrane segments also prevents erroneous targeting. These results highlight the evolution of a fine discrimination between targeting pathways that is critical for the virulence of many bacterial pathogens.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

