Evidence for intragenic recombination and selective sweep in an effector gene of Phytophthora infestans
- PMID: 30151044
- PMCID: PMC6099815
- DOI: 10.1111/eva.12629
Evidence for intragenic recombination and selective sweep in an effector gene of Phytophthora infestans
Abstract
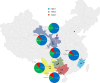
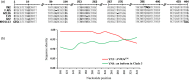
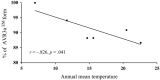
Effectors, a group of small proteins secreted by pathogens, play a critical role in the antagonistic interaction between plant hosts and pathogens through their dual functions in regulating host immune systems and pathogen infection capability. In this study, evolution in effector genes was investigated through population genetic analysis of Avr3a sequences generated from 96 Phytophthora infestans isolates collected from six locations representing a range of thermal variation and cropping systems in China. We found high genetic variation in the Avr3a gene resulting from diverse mechanisms extending beyond point mutations, frameshift, and defeated start and stop codons to intragenic recombination. A total of 51 nucleotide haplotypes encoding 38 amino acid isoforms were detected in the 96 full sequences with nucleotide diversity in the pathogen populations ranging from 0.007 to 0.023 (mean = 0.017). Although haplotype and nucleotide diversity were high, the effector gene was dominated by only three haplotypes. Evidence for a selective sweep was provided by (i) the population genetic differentiation (GST) of haplotypes being lower than the population differentiation (FST) of SSR marker loci; and (ii) negative values of Tajima's D and Fu's FS. Annual mean temperature in the collection sites was negatively correlated with the frequency of the virulent form (Avr3aEM), indicating Avr3a may be regulated by temperature. These results suggest that elevated air temperature due to global warming may hamper the development of pathogenicity traits in P. infestans and further study under confined thermal regimes may be required to confirm the hypothesis.
Keywords: Phytophthora infestans; climate change; compensatory mutation; effector genes; evolution; recombination; selective sweep; temperature dependent.
Figures

Similar articles
-
Multiple Mechanisms Drive the Evolutionary Adaptation of Phytophthora infestans Effector Avr1 to Host Resistance.J Fungi (Basel). 2021 Sep 23;7(10):789. doi: 10.3390/jof7100789. J Fungi (Basel). 2021. PMID: 34682211 Free PMC article.
-
Mutations in the signal peptide of effector gene Pi04314 contribute to the adaptive evolution of the Phytophthora infestans.BMC Ecol Evol. 2025 Mar 14;25(1):21. doi: 10.1186/s12862-025-02360-4. BMC Ecol Evol. 2025. PMID: 40082776 Free PMC article.
-
Effector Avr4 in Phytophthora infestans Escapes Host Immunity Mainly Through Early Termination.Front Microbiol. 2021 May 28;12:646062. doi: 10.3389/fmicb.2021.646062. eCollection 2021. Front Microbiol. 2021. PMID: 34122360 Free PMC article.
-
The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana.Plant J. 2006 Oct;48(2):165-76. doi: 10.1111/j.1365-313X.2006.02866.x. Epub 2006 Sep 12. Plant J. 2006. PMID: 16965554
-
Host-mediated gene silencing of a single effector gene from the potato pathogen Phytophthora infestans imparts partial resistance to late blight disease.Funct Integr Genomics. 2015 Nov;15(6):697-706. doi: 10.1007/s10142-015-0446-z. Epub 2015 Jun 16. Funct Integr Genomics. 2015. PMID: 26077032
Cited by
-
DNA methylation analysis reveals local changes in resistant and susceptible soybean lines in response to Phytophthora sansomeana.G3 (Bethesda). 2024 Oct 7;14(10):jkae191. doi: 10.1093/g3journal/jkae191. G3 (Bethesda). 2024. PMID: 39141590 Free PMC article.
-
Multiple Mechanisms Drive the Evolutionary Adaptation of Phytophthora infestans Effector Avr1 to Host Resistance.J Fungi (Basel). 2021 Sep 23;7(10):789. doi: 10.3390/jof7100789. J Fungi (Basel). 2021. PMID: 34682211 Free PMC article.
-
Lack of gene flow between Phytophthora infestans populations of two neighboring countries with the largest potato production.Evol Appl. 2019 Sep 28;13(2):318-329. doi: 10.1111/eva.12870. eCollection 2020 Feb. Evol Appl. 2019. PMID: 31993079 Free PMC article.
-
Molecular diversity, haplotype distribution and genetic variation flow of Bipolaris sorokiniana fungus causing spot blotch disease in different wheat-growing zones.J Appl Genet. 2022 Dec;63(4):793-803. doi: 10.1007/s13353-022-00716-w. Epub 2022 Aug 5. J Appl Genet. 2022. PMID: 35931929
-
Mitochondrial Genome Contributes to the Thermal Adaptation of the Oomycete Phytophthora infestans.Front Microbiol. 2022 Jun 28;13:928464. doi: 10.3389/fmicb.2022.928464. eCollection 2022. Front Microbiol. 2022. PMID: 35836411 Free PMC article.
References
-
- Allen, R. L. , Meitz, J. C. , Baumber, R. E. , Hall, S. A. , Lee, S. C. , Rose, L. E. , & Beynon, J. L. (2008). Natural variation reveals key amino acids in a downy mildew effector that alters recognition specificity by an arabidopsis resistance gene. Molecular Plant Pathology, 9, 511–523. 10.1111/j.1364-3703.2008.00481.x - DOI - PMC - PubMed
-
- Armstrong, M. R. , Whisson, S. C. , Pritchard, L. , Bos, J. I. , Venter, E. , Avrova, A. O. , … Hamlin, N. (2005). An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proceedings of the National Academy of Sciences of the United States of America, 102, 7766–7771. 10.1073/pnas.0500113102 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

