Comparative mitochondrial genomic analyses of three chemosynthetic vesicomyid clams from deep-sea habitats
- PMID: 30151147
- PMCID: PMC6106168
- DOI: 10.1002/ece3.4153
Comparative mitochondrial genomic analyses of three chemosynthetic vesicomyid clams from deep-sea habitats
Abstract
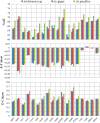
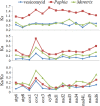
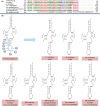
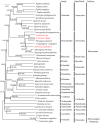
Vesicomyid clams of the subfamily Pliocardinae are among the dominant chemosymbiotic bivalves found in sulfide-rich deep-sea habitats. Plastic morphologies and present molecular data could not resolve taxonomic uncertainties. The complete mitochondrial (mt) genomes will provide more data for comparative studies on molecular phylogeny and systematics of this taxonomically uncertain group, and help to clarify generic classifications. In this study, we analyze the features and evolutionary dynamics of mt genomes from three Archivesica species (Archivesica sp., Ar. gigas and Ar. pacifica) pertaining to subfamily Pliocardinae. Sequence coverage is nearly complete for the three newly sequenced mt genomes, with only the control region and some tRNA genes missing. Gene content, base composition, and codon usage are highly conserved in these pliocardiin species. Comparative analysis revealed the vesicomyid have a relatively lower ratio of Ka/Ks, and all 13 protein-coding genes (PGCs) are under strong purifying selection with a ratio of Ka/Ks far lower than one. Minimal changes in gene arrangement among vesicomyid species are due to the translocation trnaG in Isorropodon fossajaponicum. Additional tRNA genes were detected between trnaG and nad2 in Abyssogena mariana (trnaL3), Ab. phaseoliformis (trnaS3), and Phreagena okutanii (trnaM2), and display high similarity to other pliocardiin sequences at the same location. Single base insertion in multiple sites of this location could result in new tRNA genes, suggesting a possible tRNA arising from nongeneic sequence. Phylogenetic analysis based on 12 PCGs (excluding atp8) supports the monophyly of Pliocardiinae. These nearly complete mitogenomes provide relevant data for further comparative studies on molecular phylogeny and systematics of this taxonomically uncertain group of chemosymbiotic bivalves.
Keywords: chemosymbiotic bivalves; deep‐sea; gene arrangement; mitochondrial genome; vesicomyid clams.
Figures



References
-
- Bonnaud, L. , Boucher‐Rodoni, R. , & Monnerot, M. (1994). Phylogeny of decapod cephalopods based on partial 16S rDNA nucleotide sequences. Comptes rendus de l'Academie des sciences. Serie III, Sciences de la vie, 317, 581–588. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

