A Structural Basis for 129 Xe Hyper-CEST Signal in TEM-1 β-Lactamase
- PMID: 30151973
- PMCID: PMC6611679
- DOI: 10.1002/cphc.201800624
A Structural Basis for 129 Xe Hyper-CEST Signal in TEM-1 β-Lactamase
Abstract
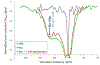
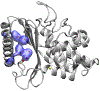
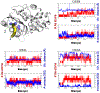
Genetically encoded (GE) contrast agents detectable by magnetic resonance imaging (MRI) enable non-invasive visualization of gene expression and cell proliferation at virtually unlimited penetration depths. Using hyperpolarized 129 Xe in combination with chemical exchange saturation transfer, an MR contrast approach known as hyper-CEST, enables ultrasensitive protein detection and biomolecular imaging. GE MRI contrast agents developed to date include nanoscale proteinaceous gas vesicles as well as the monomeric bacterial proteins TEM-1 β-lactamase (bla) and maltose binding protein (MBP). To improve understanding of hyper-CEST NMR with proteins, structural and computational studies were performed to further characterize the Xe-bla interaction. X-ray crystallography validated the location of a high-occupancy Xe binding site predicted by MD simulations, and mutagenesis experiments confirmed this Xe site as the origin of the observed CEST contrast. Structural studies and MD simulations with representative bla mutants offered additional insight regarding the relationship between local protein structure and CEST contrast.
Keywords: CEST; Xenon; contrast; hyperpolarized; magnetic resonance.
© 2019 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures




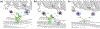

References
-
- Palmer AE, Qin Y, Park JG, McCombs JE, Trends Biotechnol. 2011, 29, 144–152. - PMC - PubMed
- Contag CH, Bachmann MH, Annu. Rev. Biomed. Eng 2002, 4, 235–260. - PubMed
- Tsien RY, Annu. Rev. Biochem 1998, 67, 509–544. - PubMed
- Lippincott-Schwartz J, Patterson GH, Science. 2003, 300, 87–91. - PubMed
- Frommer WB, Davidson MW, Campbell RE, Chem. Soc. Rev 2009, 38, 2833–2841. - PMC - PubMed
-
- Marvin JS, Borghuis BG, Tian L, Cichon J, Harnett MT, Akerboom J, Gordus A, Renninger SL, Chen T-W, Bargmann CI, Orger MB, Schreiter ER, Demb JB, Gan W-B, Hires SA, Looger LL, Nat. Methods 2013, 10, 162–70. - PMC - PubMed
- Lu Y, Stem Cells Dev. 2004, 13, 133–145. - PubMed
- Thomas CE, Ehrhardt A, Kay MA, Nat. Rev. Genet 2003, 4, 346–358. - PubMed
-
- Lyons SK, Patrick PS, Brindle KM, Cold Spring Harb. Protoc 2013, 2013, 685–699. - PubMed
- Weissleder R, Ntziachristos V, Nat. Med 2003, 9, 123–128. - PubMed
- Zhao H, Doyle TC, Coquoz O, Kalish F, Rice BW, Contag CH, J. Biomed. Opt 2011, 10, 41210. - PubMed
- Dmochowski IJ, Dmochowski JE, Oliveri P, Davidson EH and Fraser SE, Proc. Natl. Acad. Sci. U. S. A 2002, 99, 12895–12900. - PMC - PubMed
-
- Couch MJ, Blasiak B, Tomanek B, Ouriadov AV, Fox MS, Dowhos KM, Albert MS, Mol. Imaging Biol 2015, 17, 149–162. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- S10 RR029205/RR/NCRR NIH HHS/United States
- P30 GM133893/GM/NIGMS NIH HHS/United States
- P41 GM103403/GM/NIGMS NIH HHS/United States
- R01-GM097478/GF/NIH HHS/United States
- R01 GM097478/GM/NIGMS NIH HHS/United States
- R35 GM131907/GM/NIGMS NIH HHS/United States
- P41 GM111244/GM/NIGMS NIH HHS/United States
- T32-GM008275/NIH Structural Biology and Molecular Biophysics Training/International
- W81XWH-14-1-0424/Department of Defense Lung Cancer Research Progam/International
- P41 GM103393/GM/NIGMS NIH HHS/United States
- T32 GM008275/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

