Microarray expression profile of long non-coding RNAs in human lung adenocarcinoma
- PMID: 30151992
- PMCID: PMC6166069
- DOI: 10.1111/1759-7714.12845
Microarray expression profile of long non-coding RNAs in human lung adenocarcinoma
Abstract
Background: Long non-coding RNAs (lncRNAs) participate in many biological dynamics and play significant roles in gene regulation. LncRNA expression is altered in many cancers; however, the expressions and functions of lncRNA genes in lung adenocarcinoma (LAD) remain unknown.
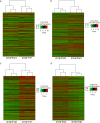
Methods: LncRNA and messenger RNA (mRNA) expression in LAD without lymphatic metastasis versus paired adjacent non-tumor (ANT) lung tissues and LAD with versus without lymphatic metastasis were analyzed using Human LncRNA Arraystar V3.0. The expression levels of four downregulated and four upregulated lncRNAs were verified using quantitative real-time PCR in cells and tissue specimens.
Results: In this study, 949 lncRNAs and 681 mRNAs had differential expression in LAD without lymphatic metastasis compared to ANT lung tissues, while 2740 lncRNAs and 1714 mRNAs were differentially expressed in LAD with lymphatic metastasis compared to LAD without lymphatic metastasis. The expression patterns of selected lncRNAs (LINC00113, AC005009.1, ARHGAP22-IT1, AC009411.1, SRGAP3-AS2, EGFEM1P, FAM66E, and HLA-F-AS1) were consistent with microarray data. Differentially expressed mRNA genes were enriched in crucial Gene Ontology terms and pathways.
Conclusion: Our results revealed differentially expressed lncRNAs in LAD, suggesting lncRNAs may be potential indicators for LAD diagnosis and therapy.
Keywords: Gene ontology; long non-coding RNA; lung adenocarcinoma; lymphatic metastasis; microarray.
© 2018 The Authors. Thoracic Cancer published by China Lung Oncology Group and John Wiley & Sons Australia, Ltd.
Figures

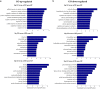
 ) Microarray and (
) Microarray and ( ) qRT‐PCR. (b) Expression of upregulated genes in referred cell lines by qRT‐PCR (
) qRT‐PCR. (b) Expression of upregulated genes in referred cell lines by qRT‐PCR ( ) LINC00113, (
) LINC00113, ( ) AC005009.1, (
) AC005009.1, ( ) ARHGAP22‐IT1, and (
) ARHGAP22‐IT1, and ( ) AC009411.1. (c) Expression of downregulated genes in referred cell lines by qRT‐PCR (
) AC009411.1. (c) Expression of downregulated genes in referred cell lines by qRT‐PCR ( ) SRGAP3‐AS2, (
) SRGAP3‐AS2, ( ) EGFEM1P, (
) EGFEM1P, ( ) FAM66E and (
) FAM66E and ( ) HLA‐F‐AS1. (Triplicate assays were performed; P < 0.05).
) HLA‐F‐AS1. (Triplicate assays were performed; P < 0.05).

References
-
- Quinn JJ, Chang HY. Unique features of long non‐coding RNA biogenesis and function. Nat Rev Genet 2016; 17: 47–62. - PubMed
-
- Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol 2016; 893: 1–19. - PubMed
-
- Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. (Published erratum appears in CA Cancer J Clin 2014; 64: 364.). CA Cancer J Clin 2014; 64: 9–29. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017; 67: 7–30. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018; 68: 7–30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

