FungiDB: An Integrated Bioinformatic Resource for Fungi and Oomycetes
- PMID: 30152809
- PMCID: PMC5872342
- DOI: 10.3390/jof4010039
FungiDB: An Integrated Bioinformatic Resource for Fungi and Oomycetes
Abstract
FungiDB (fungidb.org) is a free online resource for data mining and functional genomics analysis for fungal and oomycete species. FungiDB is part of the Eukaryotic Pathogen Genomics Database Resource (EuPathDB, eupathdb.org) platform that integrates genomic, transcriptomic, proteomic, and phenotypic datasets, and other types of data for pathogenic and nonpathogenic, free-living and parasitic organisms. FungiDB is one of the largest EuPathDB databases containing nearly 100 genomes obtained from GenBank, Aspergillus Genome Database (AspGD), The Broad Institute, Joint Genome Institute (JGI), Ensembl, and other sources. FungiDB offers a user-friendly web interface with embedded bioinformatics tools that support custom in silico experiments that leverage FungiDB-integrated data. In addition, a Galaxy-based workspace enables users to generate custom pipelines for large-scale data analysis (e.g., RNA-Seq, variant calling, etc.). This review provides an introduction to the FungiDB resources and focuses on available features, tools, and queries and how they can be used to mine data across a diverse range of integrated FungiDB datasets and records.
Keywords: Galaxy; RNA-Seq; bioinformatics; fungi; genomics; omics; pathogen; proteomics; sequence analysis; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



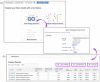


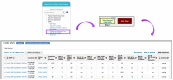
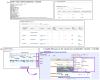

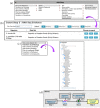







References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

