BMP4 Upregulation Is Associated with Acquired Drug Resistance and Fatty Acid Metabolism in EGFR-Mutant Non-Small-Cell Lung Cancer Cells
- PMID: 30153566
- PMCID: PMC6118096
- DOI: 10.1016/j.omtn.2018.07.016
BMP4 Upregulation Is Associated with Acquired Drug Resistance and Fatty Acid Metabolism in EGFR-Mutant Non-Small-Cell Lung Cancer Cells
Abstract
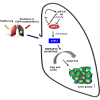
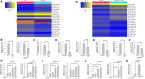
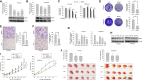
Lung cancer is the leading cause of cancer-associated deaths worldwide. In particular, non-small-cell lung cancer (NSCLC) cells harboring epidermal growth factor receptor (EGFR) mutations are associated with resistance development of EGFR tyrosine kinase inhibitor (EGFR-TKI) treatment. Recent findings suggest that bone morphogenetic proteins (BMPs) and microRNAs (miRNAs) might act as oncogenes or tumor suppressors in the tumor microenvironment. In this study, for the first time, we identified the potential roles of BMPs and miRNAs involved in EGFR-TKI resistance by analyzing datasets from a pair of parental cells and NSCLC cells with acquired EGFR-TKI resistance. BMP4 was observed to be significantly overexpressed in the EGFR-TKI-resistant cells, and its mechanism of action was strongly associated with the induction of cancer cell energy metabolism through the modulation of Acyl-CoA synthetase long-chain family member 4. In addition, miR-139-5p was observed to be significantly downregulated in the resistant NSCLC cells. The combination of miR-139-5p and yuanhuadine, a naturally derived antitumor agent, synergistically suppressed BMP4 expression in the resistant cells. We further confirmed that LDN-193189, a small molecule BMP receptor 1 inhibitor, effectively inhibited tumor growth in a xenograft nude mouse model implanted with the EFGR-TKI-resistant cells. These findings suggest a novel role of BMP4-mediated tumorigenesis in the progression of acquired drug resistance in EGFR-mutant NSCLC cells.
Keywords: Acyl-CoA synthetase long-chain family member 4; EGFR-TKI resistance; EGFR-mutant NSCLC cells; bone morphogenetic protein 4; energy metabolism; fatty acid; miR-139-5p; p53; yuanhuadine.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Engelman J.A., Settleman J. Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr. Opin. Genet. Dev. 2008;18:73–79. - PubMed
-
- Holohan C., Van Schaeybroeck S., Longley D.B., Johnston P.G. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer. 2013;13:714–726. - PubMed
-
- Nakata A., Gotoh N. Recent understanding of the molecular mechanisms for the efficacy and resistance of EGF receptor-specific tyrosine kinase inhibitors in non-small cell lung cancer. Expert Opin. Ther. Targets. 2012;16:771–781. - PubMed
-
- Sharma S.V., Bell D.W., Settleman J., Haber D.A. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer. 2007;7:169–181. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

