Escherichia coli ST131- H 22 as a Foodborne Uropathogen
- PMID: 30154256
- PMCID: PMC6113624
- DOI: 10.1128/mBio.00470-18
Escherichia coli ST131- H 22 as a Foodborne Uropathogen
Abstract
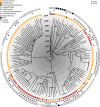
Escherichia coli sequence type 131 (ST131) has emerged rapidly to become the most prevalent extraintestinal pathogenic E. coli clones in circulation today. Previous investigations appeared to exonerate retail meat as a source of human exposure to ST131; however, these studies focused mainly on extensively multidrug-resistant ST131 strains, which typically carry allele 30 of the fimH type 1 fimbrial adhesin gene (ST131-H30). To estimate the frequency of extraintestinal human infections arising from foodborne ST131 strains without bias toward particular sublineages or phenotypes, we conducted a 1-year prospective study of E. coli from meat products and clinical cultures in Flagstaff, Arizona. We characterized all isolates by multilocus sequence typing, fimH typing, and core genome phylogenetic analyses, and we screened isolates for avian-associated ColV plasmids as an indication of poultry adaptation. E. coli was isolated from 79.8% of the 2,452 meat samples and 72.4% of the 1,735 culture-positive clinical samples. Twenty-seven meat isolates were ST131 and belonged almost exclusively (n = 25) to the ST131-H22 lineage. All but 1 of the 25 H22 meat isolates were from poultry products, and all but 2 carried poultry-associated ColV plasmids. Of the 1,188 contemporaneous human clinical E. coli isolates, 24 were ST131-H22, one-quarter of which occurred in the same high-resolution phylogenetic clades as the ST131-H22 meat isolates and carried ColV plasmids. Molecular clock analysis of an international ST131-H22 genome collection suggested that ColV plasmids have been acquired at least six times since the 1940s and that poultry-to-human transmission is not limited to the United States.IMPORTANCEE. coli ST131 is an important extraintestinal pathogen that can colonize the gastrointestinal tracts of humans and food animals. Here, we combined detection of accessory traits associated with avian adaptation (ColV plasmids) with high-resolution phylogenetics to quantify the portion of human infections caused by ST131 strains of food animal origin. Our results suggest that one ST131 sublineage-ST131-H22-has become established in poultry populations around the world and that meat may serve as a vehicle for human exposure and infection. ST131-H22 is just one of many E. coli lineages that may be transmitted from food animals to humans. Additional studies that combine detection of host-associated accessory elements with phylogenetics may allow us to quantify the total fraction of human extraintestinal infections attributable to food animal E. coli strains.
Keywords: Antibiotic resistance; ColV plasmid; Escherichia coli; ExPEC; ST131; UTI; antimicrobial resistance; foodborne; host adaptation; poultry; urinary tract infection.
Copyright © 2018 Liu et al.
Figures

References
-
- Schappert SM, Rechtsteiner EA. 2011. Ambulatory medical care utilization estimates for 2007. Vital Health Stat 13:1–38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

