The Metabolic Redox Regime of Pseudomonas putida Tunes Its Evolvability toward Novel Xenobiotic Substrates
- PMID: 30154264
- PMCID: PMC6113623
- DOI: 10.1128/mBio.01512-18
The Metabolic Redox Regime of Pseudomonas putida Tunes Its Evolvability toward Novel Xenobiotic Substrates
Abstract
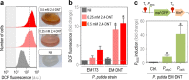
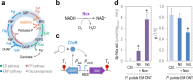
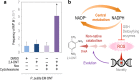
During evolution of biodegradation pathways for xenobiotic compounds involving Rieske nonheme iron oxygenases, the transition toward novel substrates is frequently associated with faulty reactions. Such events release reactive oxygen species (ROS), which are endowed with high mutagenic potential. In this study, we evaluated how the operation of the background metabolic network by an environmental bacterium may either foster or curtail the still-evolving pathway for 2,4-dinitrotoluene (2,4-DNT) catabolism. To this end, the genetically tractable strain Pseudomonas putida EM173 was implanted with the whole genetic complement necessary for the complete biodegradation of 2,4-DNT (recruited from the environmental isolate Burkholderia sp. R34). By using reporter technology and direct measurements of ROS formation, we observed that the engineered P. putida strain experienced oxidative stress when catabolizing the nitroaromatic substrate. However, the formation of ROS was neither translated into significant activation of the SOS response to DNA damage nor did it result in a mutagenic regime (unlike what has been observed in Burkholderia sp. R34, the original host of the pathway). To inspect whether the tolerance of P. putida to oxidative challenges could be traced to its characteristic reductive redox regime, we artificially altered the NAD(P)H pool by means of a water-forming, NADH-specific oxidase. Under the resulting low-NAD(P)H status, catabolism of 2,4-DNT triggered a conspicuous mutagenic and genomic diversification scenario. These results indicate that the background biochemical network of environmental bacteria ultimately determines the evolvability of metabolic pathways. Moreover, the data explain the efficacy of some bacteria (e.g., pseudomonads) to host and evolve with new catabolic routes.IMPORTANCE Some environmental bacteria evolve with new capacities for the aerobic biodegradation of chemical pollutants by adapting preexisting redox reactions to novel compounds. The process typically starts by cooption of enzymes from an available route to act on the chemical structure of the substrate-to-be. The critical bottleneck is generally the first biochemical step, and most of the selective pressure operates on reshaping the initial reaction. The interim uncoupling of the novel substrate to preexisting Rieske nonheme iron oxygenases usually results in formation of highly mutagenic ROS. In this work, we demonstrate that the background metabolic regime of the bacterium that hosts an evolving catabolic pathway (e.g., biodegradation of the xenobiotic 2,4-DNT) determines whether the cells either adopt a genetic diversification regime or a robust ROS-tolerant status. Furthermore, our results offer new perspectives to the rational design of efficient whole-cell biocatalysts, which are pursued in contemporary metabolic engineering.
Keywords: NADPH oxidases; Pseudomonas putida; biodegradation; dinitrotoluene; evolution; oxidative stress; reactive oxygen species.
Copyright © 2018 Akkaya et al.
Figures



Comment in
-
The Effect of Cellular Redox Status on the Evolvability of New Catabolic Pathways.mBio. 2018 Oct 16;9(5):e01981-18. doi: 10.1128/mBio.01981-18. mBio. 2018. PMID: 30327439 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
