Surviving host - and food relevant stresses: phenotype of L. monocytogenes strains isolated from food and clinical sources
- PMID: 30154513
- PMCID: PMC6113203
- DOI: 10.1038/s41598-018-30723-z
Surviving host - and food relevant stresses: phenotype of L. monocytogenes strains isolated from food and clinical sources
Abstract
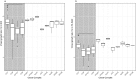
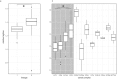
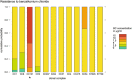
The aim of this study was to compare the phenotype of 40 strains of L. monocytogenes under food and host relevant stress conditions. The strains were chosen to represent food and clinical isolates and to be equally distributed between the most relevant clonal complexes for clinical and food isolates (CC1 and CC6 vs CC121 and CC9), plus one group of eight strains of rare clonal complexes. Human-associated CC1 had a faster maximal growth rate than the other major complexes, and the lag time of CC1 and CC6 was significantly less affected by the addition of 4% NaCl to the medium. Food-associated CC9 strains were hypohemolytic compared to other clonal complexes, and all strains found to be resistant to increased concentrations of benzalkonium chloride belonged to CC121 and were positive for Tn6188 carrying the qacH gene. Lactic acid affected the survival of L. monocytogenes more than HCl, and there was a distinct, strain specific pattern of acid tolerant and sensitive strains. Strains from CC6 and human clinical isolates are less resilient under acid stress than those from other complexes and from food. One strain isolated from a human patient exhibited significant growth defects across all conditions.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- de Valk H, et al. Surveillance of Listeria infections in Europe. Euro Surveill. Bull. Eur. sur les Mal. Transm. = Eur. Commun. Dis. Bull. 2005;10:251–255. - PubMed
-
- Kovacevic J, Arguedas-Villa C, Wozniak A, Tasara T, Allen KJ. Examination of food chain-derived Listeria monocytogenes strains of different serotypes reveals considerable diversity in inlA genotypes, mutability, and adaptation to cold temperatures. Appl. Environ. Microbiol. 2013;79:1915–1922. doi: 10.1128/AEM.03341-12. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

