Phosphorylation promotes binding affinity of Rap-Raf complex by allosteric modulation of switch loop dynamics
- PMID: 30154518
- PMCID: PMC6113251
- DOI: 10.1038/s41598-018-31234-7
Phosphorylation promotes binding affinity of Rap-Raf complex by allosteric modulation of switch loop dynamics
Abstract
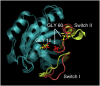
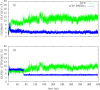
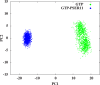
The effects of phosphorylation of a serine residue on the structural and dynamic properties of Ras-like protein, Rap, and its interactions with effector protein Ras binding domain (RBD) of Raf kinase, in the presence of GTP, are investigated via molecular dynamics simulations. The simulations show that phosphorylation significantly effects the dynamics of functional loops of Rap which participate in the stability of the complex with effector proteins. The effects of phosphorylation on Rap are significant and detailed conformational analysis suggest that the Rap protein, when phosphorylated and with GTP ligand, samples different conformational space as compared to non-phosphorylated protein. In addition, phosphorylation of SER11 opens up a new cavity in the Rap protein which can be further explored for possible drug interactions. Residue network analysis shows that the phosphorylation of Rap results in a community spanning both Rap and RBD and strongly suggests transmission of allosteric effects of local alterations in Rap to distal regions of RBD, potentially affecting the downstream signalling. Binding free energy calculations suggest that phosphorylation of SER11 residue increases the binding between Rap and Raf corroborating the network analysis results. The increased binding of the Rap-Raf complex can have cascading effects along the signalling pathways where availability of Raf can influence the oncogenic effects of Ras proteins. These simulations underscore the importance of post translational modifications like phosphorylation on the functional dynamics in proteins and can be an alternative to drug-targeting, especially in notoriously undruggable oncoproteins belonging to Ras-like GTPase family.
Conflict of interest statement
The authors declare no competing interests.
Figures
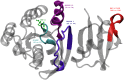






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

