Releasable and traceless PEGylation of arginine-rich antimicrobial peptides
- PMID: 30155213
- PMCID: PMC6094173
- DOI: 10.1039/c7sc00770a
Releasable and traceless PEGylation of arginine-rich antimicrobial peptides
Abstract
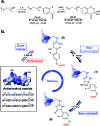
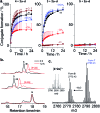
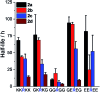
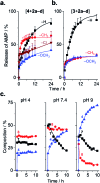
Arginine-rich antimicrobial peptides (AMPs) are emerging therapeutics of interest. However, their applicability is limited by their short circulation half-life, caused in part by their small size and digestion by blood proteases. This study reports a strategy to temporarily mask arginine residues within AMPs with methoxy poly(ethylene glycol). Based on the reagent used, release of AMPs occurred in hours to days in a completely traceless fashion. In vitro, conjugates were insensitive to serum proteases, and released native AMP with full in vitro bioactivity. This strategy is thus highly relevant and should be adaptable to the entire family of arginine-rich AMPs. It may potentially be used to improve AMP-therapies by providing a more steady concentration of AMP in the blood after a single injection, avoiding toxic effects at high AMP doses, and reducing the number of doses required over the treatment duration.
Figures
References
-
- Hancock R. E. W., Sahl H.-G. Nat. Biotechnol. 2006;24:1551–1557. - PubMed
-
- Boman H. G. J. Intern. Med. 2003;254:197–215. - PubMed
-
- Fjell C. D., Hiss J. A., Hancock R. E. W., Schneider G. Nat. Rev. Drug Discovery. 2012;11:37–51. - PubMed
-
- Chan D. I., Prenner E. J., Vogel H. J. Biochim. Biophys. Acta. 2006;1758:1184–1202. - PubMed
-
- Veronese F. M. Biomaterials. 2001;22:405–417. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

