Large-scale comparison of machine learning methods for drug target prediction on ChEMBL
- PMID: 30155234
- PMCID: PMC6011237
- DOI: 10.1039/c8sc00148k
Large-scale comparison of machine learning methods for drug target prediction on ChEMBL
Abstract
Deep learning is currently the most successful machine learning technique in a wide range of application areas and has recently been applied successfully in drug discovery research to predict potential drug targets and to screen for active molecules. However, due to (1) the lack of large-scale studies, (2) the compound series bias that is characteristic of drug discovery datasets and (3) the hyperparameter selection bias that comes with the high number of potential deep learning architectures, it remains unclear whether deep learning can indeed outperform existing computational methods in drug discovery tasks. We therefore assessed the performance of several deep learning methods on a large-scale drug discovery dataset and compared the results with those of other machine learning and target prediction methods. To avoid potential biases from hyperparameter selection or compound series, we used a nested cluster-cross-validation strategy. We found (1) that deep learning methods significantly outperform all competing methods and (2) that the predictive performance of deep learning is in many cases comparable to that of tests performed in wet labs (i.e., in vitro assays).
Figures
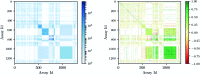



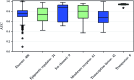

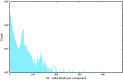
References
-
- Molina D. M., Jafari R., Ignatushchenko M., Seki T., Larsson E. A., Dan C., Sreekumar L., Cao Y., Nordlund P. Science. 2013;341:84–87. - PubMed
-
- Huang R., Xia M., Nguyen D.-T., Zhao T., Sakamuru S., Zhao J., Shahane S. A., Rossoshek A., Simeonov A. Front. Environ. Sci. Eng. 2016;3:85.
-
- Ma J., Sheridan R. P., Liaw A., Dahl G. E., Svetnik V. J. Chem. Inf. Model. 2015;55:263–274. - PubMed
-
- Mayr A., Klambauer G., Unterthiner T., Hochreiter S. Front. Environ. Sci. Eng. 2016;3:80.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

