Computational Structural Biology of S-nitrosylation of Cancer Targets
- PMID: 30155439
- PMCID: PMC6102371
- DOI: 10.3389/fonc.2018.00272
Computational Structural Biology of S-nitrosylation of Cancer Targets
Abstract
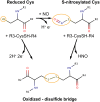
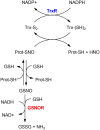
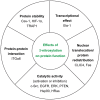
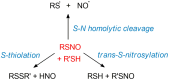
Nitric oxide (NO) plays an essential role in redox signaling in normal and pathological cellular conditions. In particular, it is well known to react in vivo with cysteines by the so-called S-nitrosylation reaction. S-nitrosylation is a selective and reversible post-translational modification that exerts a myriad of different effects, such as the modulation of protein conformation, activity, stability, and biological interaction networks. We have appreciated, over the last years, the role of S-nitrosylation in normal and disease conditions. In this context, structural and computational studies can help to dissect the complex and multifaceted role of this redox post-translational modification. In this review article, we summarized the current state-of-the-art on the mechanism of S-nitrosylation, along with the structural and computational studies that have helped to unveil its effects and biological roles. We also discussed the need to move new steps forward especially in the direction of employing computational structural biology to address the molecular and atomistic details of S-nitrosylation. Indeed, this redox modification has been so far an underappreciated redox post-translational modification by the computational biochemistry community. In our review, we primarily focus on S-nitrosylated proteins that are attractive cancer targets due to the emerging relevance of this redox modification in a cancer setting.
Keywords: (de)nitrosylating enzymes; S-nitrosylation; cysteine; molecular dynamics simulations; redox cancer biology; redox modifications.
Figures
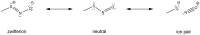
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

