A novel panel of short mononucleotide repeats linked to informative polymorphisms enabling effective high volume low cost discrimination between mismatch repair deficient and proficient tumours
- PMID: 30157243
- PMCID: PMC6114912
- DOI: 10.1371/journal.pone.0203052
A novel panel of short mononucleotide repeats linked to informative polymorphisms enabling effective high volume low cost discrimination between mismatch repair deficient and proficient tumours
Abstract
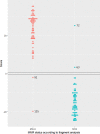
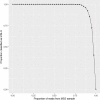
Somatic mutations in mononucleotide repeats are commonly used to assess the mismatch repair status of tumours. Current tests focus on repeats with a length above 15bp, which tend to be somatically more unstable than shorter ones. These longer repeats also have a substantially higher PCR error rate, and tests that use capillary electrophoresis for fragment size analysis often require expert interpretation. In this communication, we present a panel of 17 short repeats (length 7-12bp) for sequence-based microsatellite instability (MSI) testing. Using a simple scoring procedure that incorporates the allelic distribution of the mutant repeats, and analysis of two cohort of tumours totalling 209 samples, we show that this panel is able to discriminate between MMR proficient and deficient tumours, even when constitutional DNA is not available. In the training cohort, the method achieved 100% concordance with fragment analysis, while in the testing cohort, 4 discordant samples were observed (corresponding to 97% concordance). Of these, 2 showed discrepancies between fragment analysis and immunohistochemistry and one was reclassified after re-testing using fragment analysis. These results indicate that our approach offers the option of a reliable, scalable routine test for MSI.
Conflict of interest statement
JB is medical director and chairman of the board at QuantuMDx Ltd. LR and HS received financial support in the form of salaries from QuantuMDx Ltd. JB, MSJ, MSK, LR and GA hold a patent covering the assay markers (Patent ID: PCT/GB2017/052488). This does not alter our adherence to PLOS ONE policies on sharing data and materials. Other authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

