Transcriptome dynamics associated with resistance and susceptibility against fusarium head blight in four wheat genotypes
- PMID: 30157778
- PMCID: PMC6116500
- DOI: 10.1186/s12864-018-5012-3
Transcriptome dynamics associated with resistance and susceptibility against fusarium head blight in four wheat genotypes
Abstract
Background: Fusarium head blight (FHB) of wheat in North America is caused mostly by the fungal pathogen Fusarium graminearum (Fg). Upon exposure to Fg, wheat initiates a series of cellular responses involving massive transcriptional reprogramming. In this study, we analyzed transcriptomics data of four wheat genotypes (Nyubai, Wuhan 1, HC374, and Shaw), at 2 and 4 days post inoculation (dpi) with Fg, using RNA-seq technology.
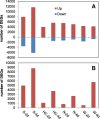
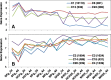
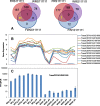
Results: A total of 37,772 differentially expressed genes (DEGs) were identified, 28,961 from wheat and 8811 from the pathogen. The susceptible genotype Shaw exhibited the highest number of host and pathogen DEGs, including 2270 DEGs associating with FHB susceptibility. Protein serine/threonine kinases and LRR-RK were associated with susceptibility at 2 dpi, while several ethylene-responsive, WRKY, Myb, bZIP and NAC-domain containing transcription factors were associated with susceptibility at 4 dpi. In the three resistant genotypes, 220 DEGs were associated with resistance. Glutathione S-transferase (GST), membrane proteins and distinct LRR-RKs were associated with FHB resistance across the three genotypes. Genes with unique, high up-regulation by Fg in Wuhan 1 were mostly transiently expressed at 2 dpi, while many defense-associated genes were up-regulated at both 2 and 4 dpi in Nyubai; the majority of unique genes up-regulated in HC374 were detected at 4 dpi only. In the pathogen, most genes showed increased expression between 2 and 4 dpi in all genotypes, with stronger levels in the susceptible host; however two pectate lyases and a hydrolase were expressed higher at 2 dpi, and acetyltransferase activity was highly enriched at 4 dpi.
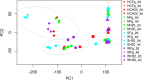
Conclusions: There was an early up-regulation of LRR-RKs, different between susceptible and resistant genotypes; subsequently, distinct sets of genes associated with defense response were up-regulated. Differences in expression profiles among the resistant genotypes indicate genotype-specific defense mechanisms. This study also shows a greater resemblance in transcriptomics of HC374 to Nyubai, consistent with their sharing of two FHB resistance QTLs on 3BS and 5AS, compared to Wuhan 1 which carries one QTL on 2DL in common with HC374.
Keywords: Differentially expressed genes; Fusarium graminearum; Fusarium head blight; Pathogenesis; Plant defense; RNA-seq; Triticum aestivum.
Conflict of interest statement
Ethics approval
Ethics approval is not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures









References
-
- Mesterházy Á, Bartók T, Mirocha CG, Komoróczy R. Nature of wheat resistance to fusarium head blight and the role of deoxynivalenol for breeding. Plant Breed. 1999;118:97–110. doi: 10.1046/j.1439-0523.1999.118002097.x. - DOI
-
- Buerstmayr H, Ban T, Anderson JA. QTL mapping and marker-assisted selection for fusarium head blight resistance in wheat: a review. Plant Breed. 2009;128:1–26. doi: 10.1111/j.1439-0523.2008.01550.x. - DOI
MeSH terms
Substances
Grants and funding
- project J-000412/Canadian Agriculture and Agri-Food Growing Forward 2 program
- projects J-000008 and J-001580/Genomics Research and Development Initiative
- National Wheat Improvement Program/Western Grains Research Foundation and Agriculture and Agri-Food Canada
- "Canadian Wheat Improvement" project A1-011652/National Research Council Canada
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

