FDHE-IW: A Fast Approach for Detecting High-Order Epistasis in Genome-Wide Case-Control Studies
- PMID: 30158504
- PMCID: PMC6162554
- DOI: 10.3390/genes9090435
FDHE-IW: A Fast Approach for Detecting High-Order Epistasis in Genome-Wide Case-Control Studies
Abstract
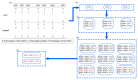
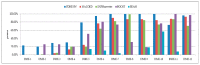
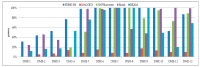
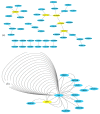
Detecting high-order epistasis in genome-wide association studies (GWASs) is of importance when characterizing complex human diseases. However, the enormous numbers of possible single-nucleotide polymorphism (SNP) combinations and the diversity among diseases presents a significant computational challenge. Herein, a fast method for detecting high-order epistasis based on an interaction weight (FDHE-IW) method is evaluated in the detection of SNP combinations associated with disease. First, the symmetrical uncertainty (SU) value for each SNP is calculated. Then, the top-k SNPs are isolated as guiders to identify 2-way SNP combinations with significant interaction weight values. Next, a forward search is employed to detect high-order SNP combinations with significant interaction weight values as candidates. Finally, the findings were statistically evaluated using a G-test to isolate true positives. The developed algorithm was used to evaluate 12 simulated datasets and an age-related macular degeneration (AMD) dataset and was shown to perform robustly in the detection of some high-order disease-causing models.
Keywords: Single-nucleotide polymorphism; high-order epistasis; interaction weight.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
HiSSI: high-order SNP-SNP interactions detection based on efficient significant pattern and differential evolution.BMC Med Genomics. 2019 Dec 30;12(Suppl 7):139. doi: 10.1186/s12920-019-0584-6. BMC Med Genomics. 2019. PMID: 31888641 Free PMC article.
-
A Novel Multitasking Ant Colony Optimization Method for Detecting Multiorder SNP Interactions.Interdiscip Sci. 2022 Dec;14(4):814-832. doi: 10.1007/s12539-022-00530-2. Epub 2022 Jul 5. Interdiscip Sci. 2022. PMID: 35788965
-
Niche harmony search algorithm for detecting complex disease associated high-order SNP combinations.Sci Rep. 2017 Sep 14;7(1):11529. doi: 10.1038/s41598-017-11064-9. Sci Rep. 2017. PMID: 28912584 Free PMC article.
-
Genotype distribution-based inference of collective effects in genome-wide association studies: insights to age-related macular degeneration disease mechanism.BMC Genomics. 2016 Aug 30;17(1):695. doi: 10.1186/s12864-016-2871-3. BMC Genomics. 2016. PMID: 27576376 Free PMC article.
-
ClusterMI: Detecting High-Order SNP Interactions Based on Clustering and Mutual Information.Int J Mol Sci. 2018 Aug 2;19(8):2267. doi: 10.3390/ijms19082267. Int J Mol Sci. 2018. PMID: 30072632 Free PMC article.
Cited by
-
SEEI: spherical evolution with feedback mechanism for identifying epistatic interactions.BMC Genomics. 2024 May 13;25(1):462. doi: 10.1186/s12864-024-10373-4. BMC Genomics. 2024. PMID: 38735952 Free PMC article.
-
A Novel Detection Method for High-Order SNP Epistatic Interactions Based on Explicit-Encoding-Based Multitasking Harmony Search.Interdiscip Sci. 2024 Sep;16(3):688-711. doi: 10.1007/s12539-024-00621-2. Epub 2024 Jul 2. Interdiscip Sci. 2024. PMID: 38954231
-
MDSN: A Module Detection Method for Identifying High-Order Epistatic Interactions.Genes (Basel). 2022 Dec 18;13(12):2403. doi: 10.3390/genes13122403. Genes (Basel). 2022. PMID: 36553670 Free PMC article.
-
EpiReSIM: A Resampling Method of Epistatic Model without Marginal Effects Using Under-Determined System of Equations.Genes (Basel). 2022 Dec 4;13(12):2286. doi: 10.3390/genes13122286. Genes (Basel). 2022. PMID: 36553553 Free PMC article.
-
MIDESP: Mutual Information-Based Detection of Epistatic SNP Pairs for Qualitative and Quantitative Phenotypes.Biology (Basel). 2021 Sep 16;10(9):921. doi: 10.3390/biology10090921. Biology (Basel). 2021. PMID: 34571798 Free PMC article.
References
-
- Jiang R. Gene-gene interaction. In: Gellman M.D., Turner J.R., editors. Encyclopedia of Behavioral Medicine. Springer; New York, NY, USA: 2013. pp. 841–842.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

