EMT- and stroma-related gene expression and resistance to PD-1 blockade in urothelial cancer
- PMID: 30158554
- PMCID: PMC6115401
- DOI: 10.1038/s41467-018-05992-x
EMT- and stroma-related gene expression and resistance to PD-1 blockade in urothelial cancer
Abstract
Cancers infiltrated with T-cells are associated with a higher likelihood of response to PD-1/PD-L1 blockade. Counterintuitively, a correlation between epithelial-mesenchymal transition (EMT)-related gene expression and T-cell infiltration has been observed across tumor types. Here we demonstrate, using The Cancer Genome Atlas (TCGA) urothelial cancer dataset, that although a gene expression-based measure of infiltrating T-cell abundance and EMT-related gene expression are positively correlated, these signatures convey disparate prognostic information. We further demonstrate that non-hematopoietic stromal cells are a major source of EMT-related gene expression in bulk urothelial cancer transcriptomes. Finally, using a cohort of patients with metastatic urothelial cancer treated with a PD-1 inhibitor, nivolumab, we demonstrate that in patients with T-cell infiltrated tumors, higher EMT/stroma-related gene expression is associated with lower response rates and shorter progression-free and overall survival. Together, our findings suggest a stroma-mediated source of immune resistance in urothelial cancer and provide rationale for co-targeting PD-1 and stromal elements.
Conflict of interest statement
M.D.G. has served as a consultant and has received research funding from Bristol-Myers Squibb. A.S., P.M.S., S.D.C., and A.A. are employees of Bristol-Myers Squibb. The remaining authors declare no competing interests.
Figures
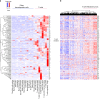
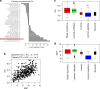





Comment in
-
Re: EMT- and Stroma-Related Gene Expression and Resistance to PD-1 Blockade in Urothelial Cancer.J Urol. 2019 Sep;202(3):458. doi: 10.1097/01.JU.0000569196.28580.f2. Epub 2019 Aug 8. J Urol. 2019. PMID: 31180289 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

